25 -- 29 August notes
2014-08-25
ApGxL
- Library concentration not adequate for Illumina sequencing
- Ran PCR of Ovation 3’ DGE library to get adequate DNA
- Used KAPA library quant primer mix as I know it worked for qPCR
- Evaluating the number of cycles necessary for adequate amplification
- 4, 6, 8, 10 cycles
- 0.25 ul of library added to each PCR (diluted in 3.75 ul T10E0.1)
- included KAPA Illumina Standards 1-3 as controls
Results
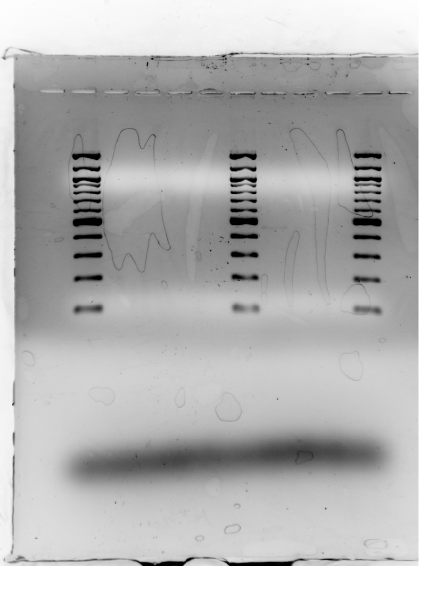
Nothing! Ladder completely clear but no bands…maybe a light light band for Standard 1
ApTranscriptome
Manuscript prep
2014-08-26
ApGxL
- Repeated yesterdays’ attempt at PCR amplification, with changes
- started with 1 ul DNA in each sample
- 12, 16, 20 and 24 cycles
- for fun, ran yesterday’s PCR samples for full 24 cycles
Results
Amplification! All samples and standards show up clearly.
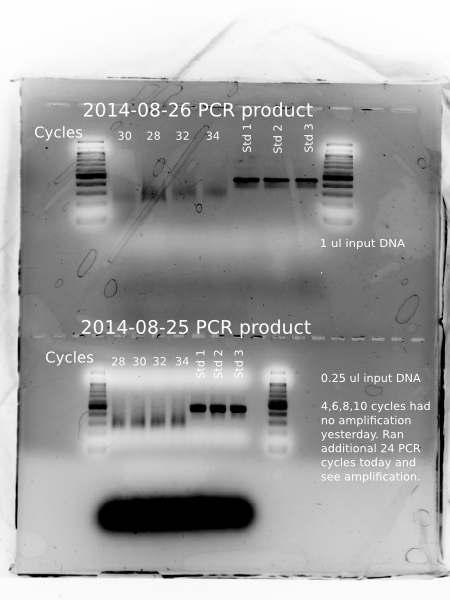
- today’s samples (12 – 24 cycles), 16 is clearly darker than 12 cycles, but 20 and 24 do not appear darker.
- yesterday’s samples (4 – 10 + 24 cycles) look similarly intense
- size of amplified fragment 300-400bp, which is consistent with expectations given 122bp of adapter sequence added to insert per NuGen
appears to be slight shift towards larger fragment sizes with increasing PCR cycles
Purified PCR product using MinElute
- Ran KAPA qPCR on C16 and C20 (1:1000 and 1:2000 dilutions)
Beautiful results! Standard r2=0.99
| Well | Sample.Name | Quantity | Cт | Dilution |
|---|---|---|---|---|
| A1 | Std1 | 20 | 9.354 | 0.001 |
| A2 | Std2 | 2 | 12.62 | 5e-04 |
| A3 | Std3 | 0.2 | 16.9 | 0.001 |
| A4 | Std4 | 0.02 | 20.14 | 5e-04 |
| A5 | Std5 | 0.002 | 22.4 | 0.001 |
| A6 | Std6 | 2e-04 | 26.37 | 5e-04 |
| A7 | C16 | 121.2 | 6.597 | 0.001 |
| A8 | C16 | 50.67 | 7.917 | 5e-04 |
| A9 | C20 | 263 | 5.425 | 0.001 |
| A10 | C20 | 98.94 | 6.904 | 5e-04 |
| B1 | Std1 | 20 | 8.674 | 0.001 |
| B2 | Std2 | 2 | 13.05 | 5e-04 |
| B3 | Std3 | 0.2 | 16.62 | 0.001 |
| B4 | Std4 | 0.02 | 19.89 | 5e-04 |
| B5 | Std5 | 0.002 | 23.22 | 0.001 |
| B6 | Std6 | 2e-04 | 27.17 | 5e-04 |
| B7 | C16 | 134.5 | 6.439 | 0.001 |
| B8 | C16 | 58.15 | 7.709 | 5e-04 |
| B9 | C20 | 238.5 | 5.573 | 0.001 |
| B10 | C20 | 134.5 | 6.439 | 5e-04 |
| C7 | C16 | 106 | 6.8 | 0.001 |
| C8 | C16 | 71.55 | 7.395 | 5e-04 |
| C9 | C20 | 255 | 5.472 | 0.001 |
| C10 | C20 | 144.7 | 6.329 | 5e-04 |
Quantification of DNA
| Sample.Name | KAPA.pM | KAPA.nM |
|---|---|---|
| C16 | 181,417 | 181.42 |
| C20 | 379,907 | 379.91 |
200x increase over un-amplified library! More than enough for sequencing!
2014-08-27
ApGxL
To try and limit the number of PCR cycles, and thus potential for bias, in library prep, did a 3rd round of PCR. Used 4 ul of DNA template at 6, 8, 10, 11 (missed 10) cycles.
KAPA library quant
| Well | Sample.Name | Quantity | Cт |
|---|---|---|---|
| A1 | Std1 | 20 | 8.851 |
| A2 | Std2 | 2 | 12.93 |
| A3 | Std3 | 0.2 | 16.41 |
| A4 | Std4 | 0.02 | 19.48 |
| A5 | Std5 | 0.002 | 22.77 |
| A6 | Std6 | 2e-04 | 25.67 |
| A7 | C6 | 4.674 | 11.4 |
| A8 | C6 | 2.009 | 12.65 |
| A9 | C8 | 13.59 | 9.824 |
| A10 | C8 | 4.726 | 11.38 |
| A11 | C11 | 49.46 | 7.915 |
| A12 | C11 | 21.25 | 9.163 |
| B1 | Std1 | 20 | 8.523 |
| B2 | Std2 | 2 | 13.04 |
| B3 | Std3 | 0.2 | 16.57 |
| B4 | Std4 | 0.02 | 19.61 |
| B5 | Std5 | 0.002 | 23.07 |
| B6 | Std6 | 2e-04 | 26.2 |
| B7 | C6 | 4.226 | 11.55 |
| B8 | C6 | 2.212 | 12.51 |
| B9 | C8 | 12.24 | 9.979 |
| B10 | C8 | 6.264 | 10.97 |
| B11 | C11 | 52.57 | 7.825 |
| B12 | C11 | 21.6 | 9.139 |
| C7 | C6 | 4.002 | 11.63 |
| C8 | C6 | 2.806 | 12.16 |
| C9 | C8 | 16.5 | 9.538 |
| C10 | C8 | 6.983 | 10.81 |
| C11 | C11 | 50.42 | 7.886 |
| C12 | C11 | 26.46 | 8.84 |
Library quantification for each PCR sample
| Sample.Name | KAPA.pM | KAPA.nM |
|---|---|---|
| C6 | 6770 | 6.77 |
| C8 | 19656 | 19.66 |
| C11 | 73091 | 73.09 |
Took sample all sample C8 and sent for sequencing - library CC02!
Also dropped off the second ApPopGen ddRADseq library prepared by SHC - diluted to 10nm in 20ul - library CC03.
Created spreadsheet to keep track of all sequencing libraries for Climate Cascade project. Each library will be labeled ‘CCXX’ with XX being digits running sequentially from 01. Saved this spreadsheet in Dropbox /Dropbox/WarmAntDimensions/Genomics/UVM-AGTC-sequencing.csv for collaborators.
- Feed Grace’s Pogos.
ApTranscriptome
Manuscript…still waiting for workstation to come back online for updates to MS.
## 2014-08-28
ApTranscriptome
Writing…
Reading
Gerstein, M.B., Rozowsky, J., Yan, K.-K., Wang, D., Cheng, C., Brown, J.B., Davis, C.A., Hillier, L., Sisu, C., Li, J.J., et al. (2014). Comparative analysis of the transcriptome across distant species. Nature 512, 445–448.
Karasov, T.L., Kniskern, J.M., Gao, L., DeYoung, B.J., Ding, J., Dubiella, U., Lastra, R.O., Nallu, S., Roux, F., Innes, R.W., et al. (2014). The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature 512, 436–440.
Muerdter, F., and Stark, A. (2014). Genomics: Hiding in plain sight. Nature 512, 374–375.
- AN practice talk
- Using R with Amazon RDS
