ddRADseq success
Finally, success with the ddRADseq protocol!
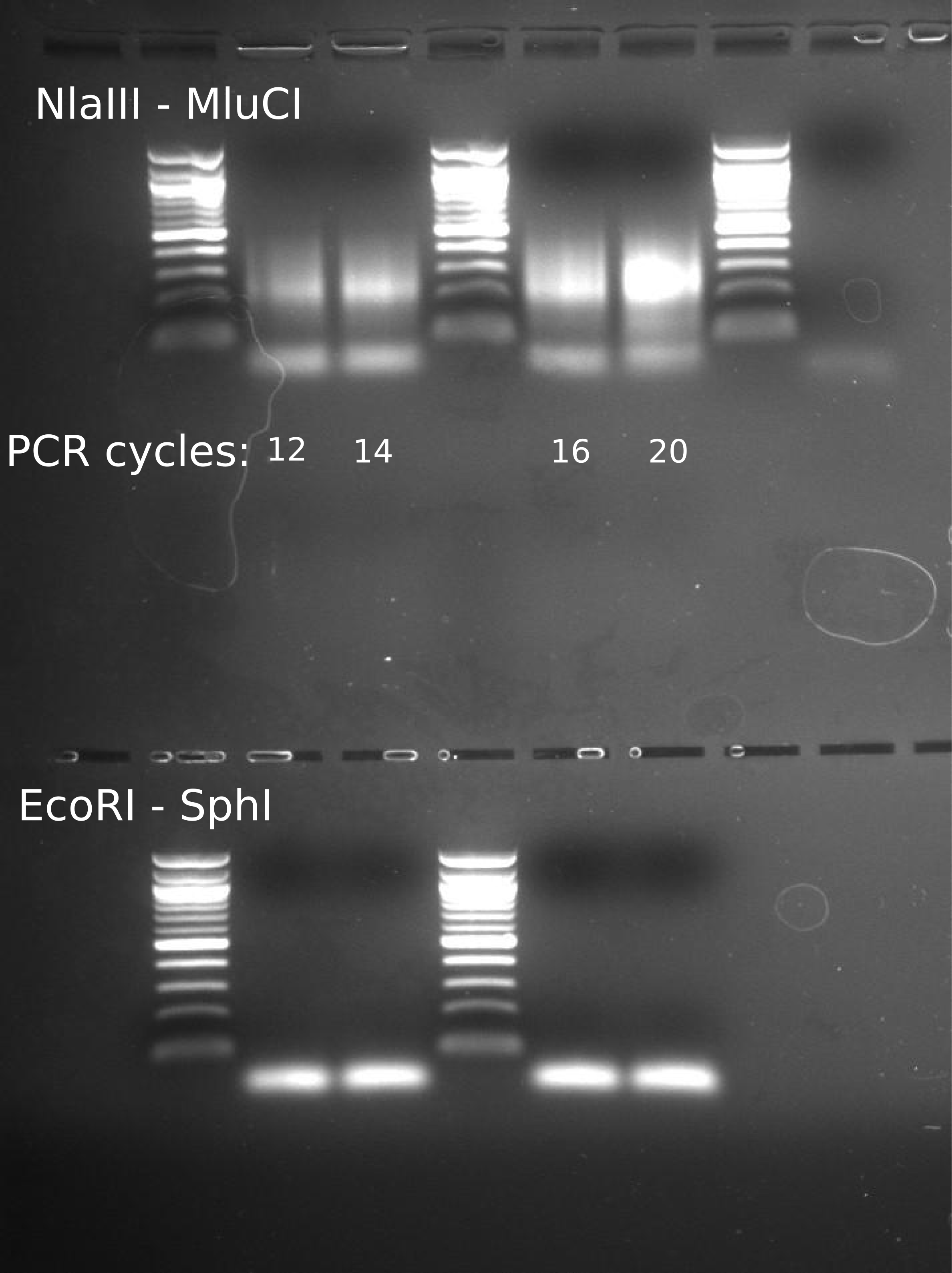
Image clearly shows that for the NlaIII - MluCI digestion and ligation, increased amplification with more cycles. This demonstrates that ligation of the P1 and P2 adapters occurred correctly.
For this de-bugging round, I started with a new P2 primer (ordered from IDT) that was not biotinylated. I’m not sure if this explains success as I suspect it may simply have to do with better lab technique, especially during the purification steps, so more DNA is retained at each step.
