![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-00-R1_val_1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 16351272 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 40 |
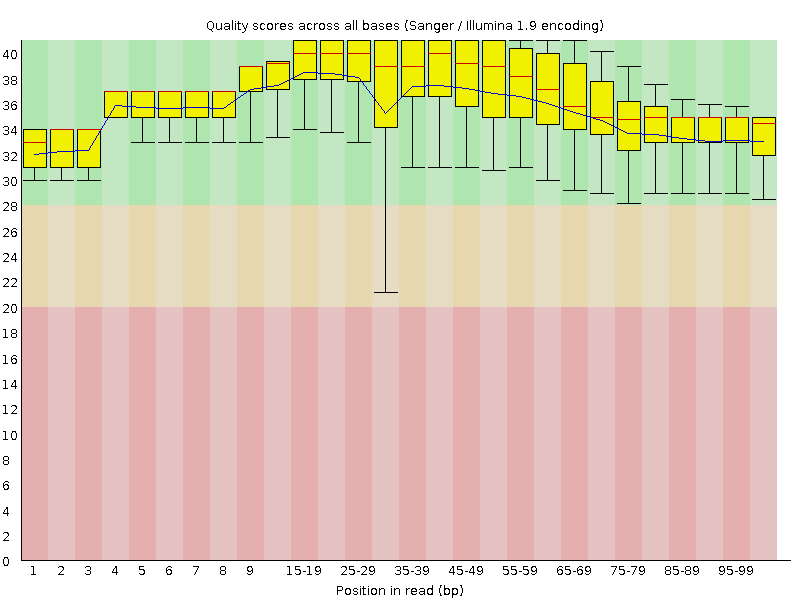
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
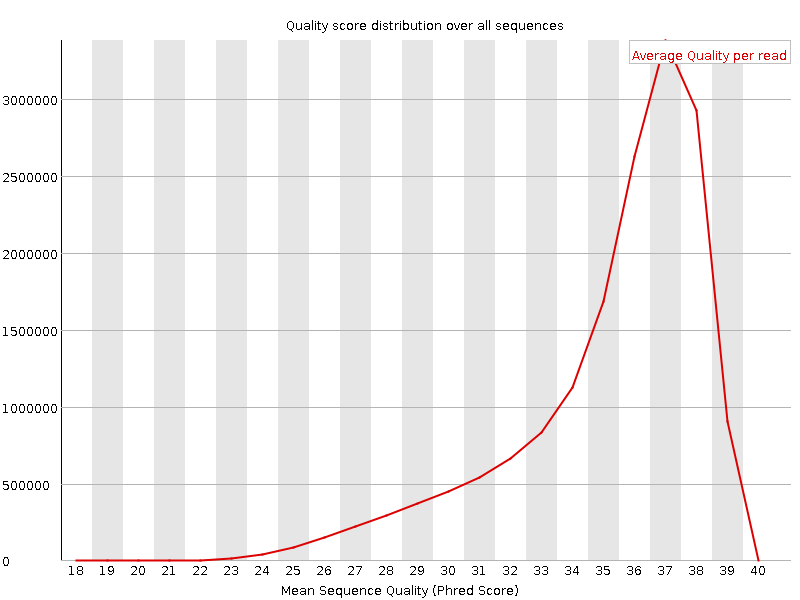
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
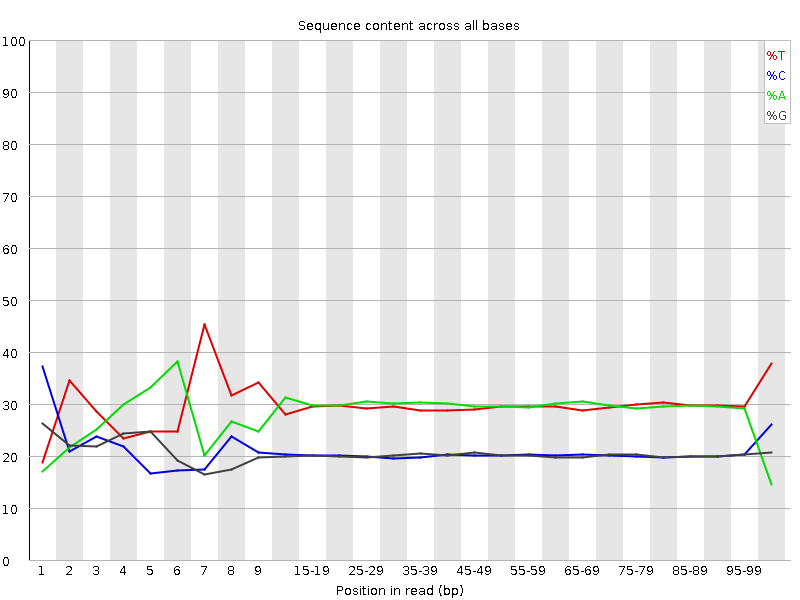
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
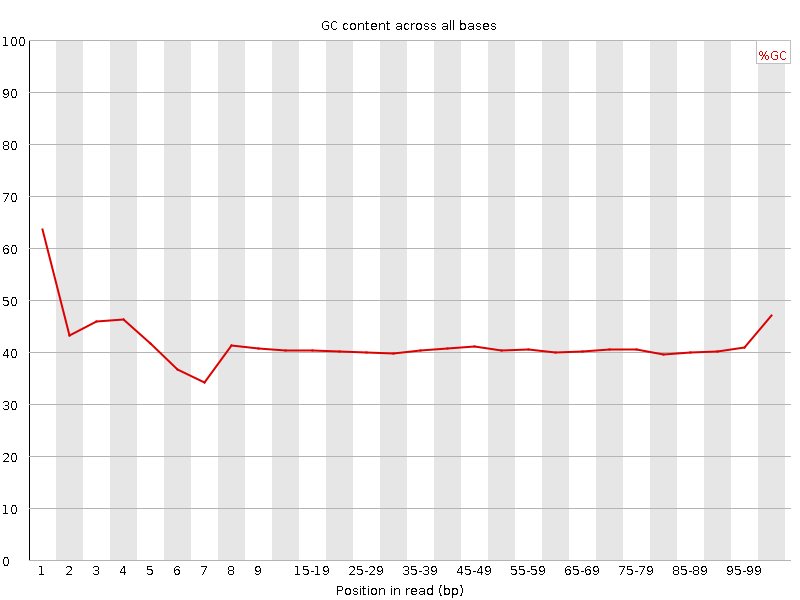
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
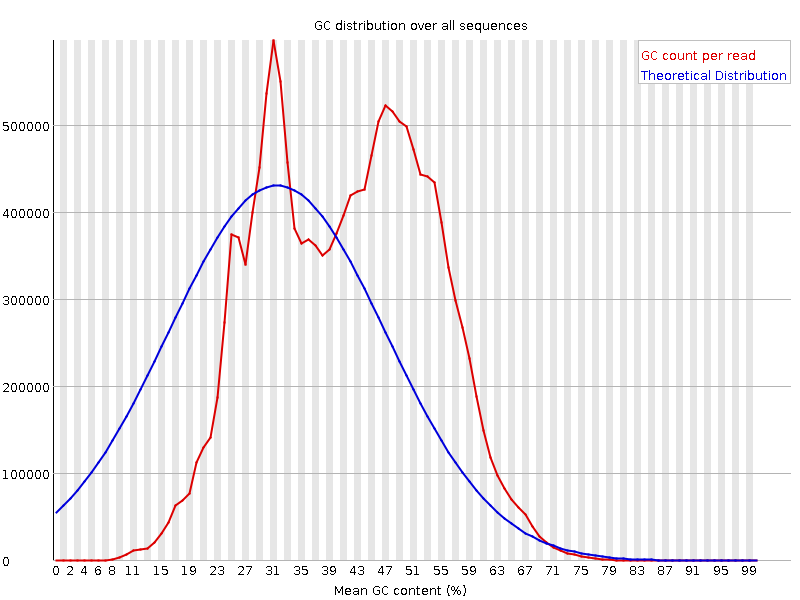
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content

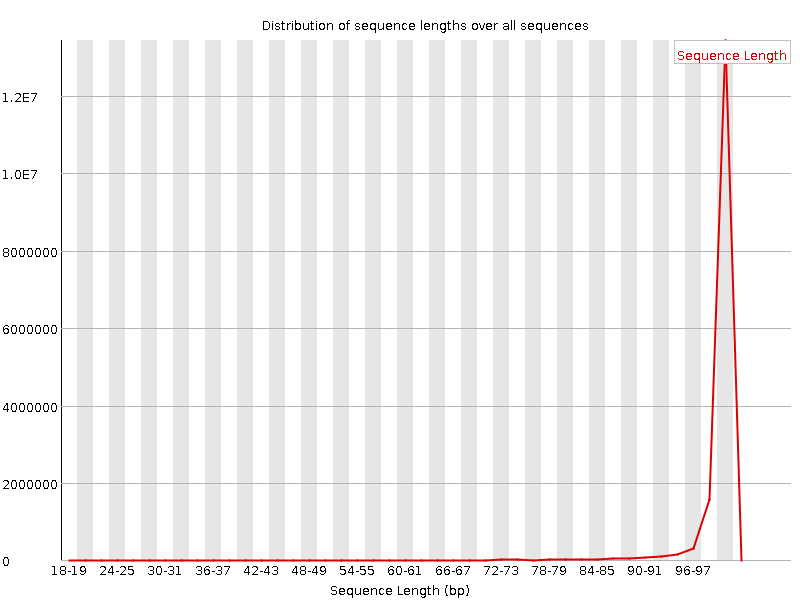
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
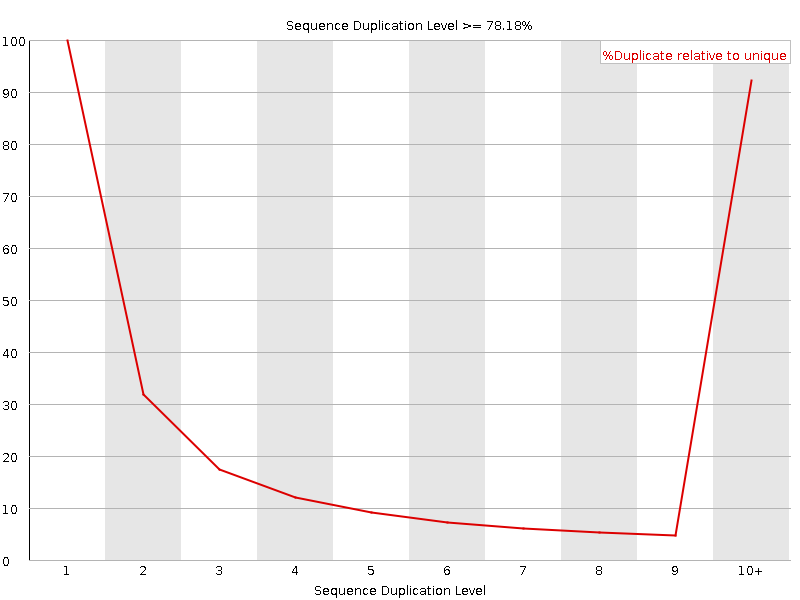
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 52213 | 0.31932072318288146 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 45712 | 0.27956234842157845 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 40548 | 0.24798070755596263 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 33350 | 0.20395966748030367 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 25904 | 0.1584219258293789 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 25523 | 0.1560918318770552 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 24891 | 0.15222668915299067 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 20978 | 0.1282958292174456 | No Hit |
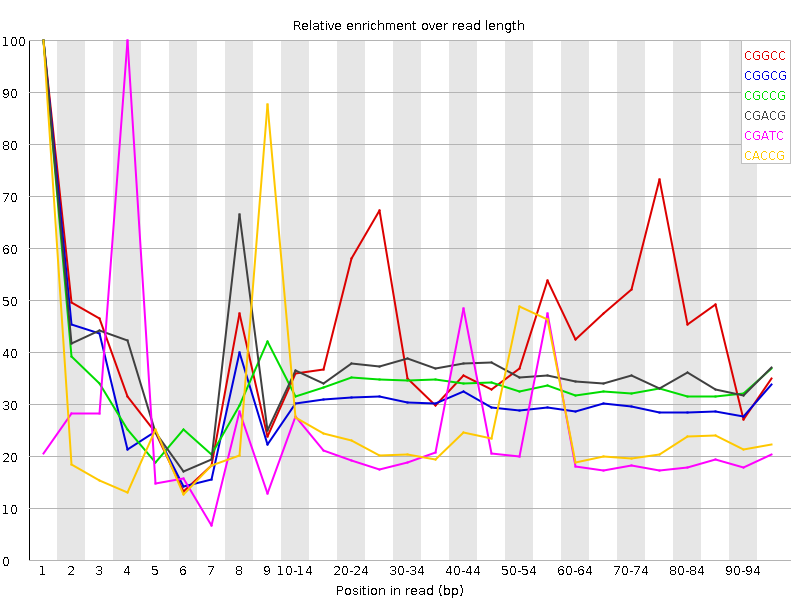
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1375125 | 2.5035539 | 5.7012625 | 1 |
| CGGCG | 1315835 | 2.412451 | 7.875865 | 1 |
| CGCCG | 1284405 | 2.3383894 | 6.93492 | 1 |
| CGACG | 1625945 | 2.057555 | 5.6467323 | 1 |
| CGATC | 2364665 | 2.0535886 | 8.755167 | 4 |
| CACCG | 1580935 | 1.9866318 | 7.626138 | 1 |
| CCTCG | 1554050 | 1.9416815 | 5.3366427 | 1 |
| CGCGG | 977220 | 1.7916346 | 5.626359 | 1 |
| CCGCG | 978070 | 1.7806752 | 5.535573 | 1 |
| CCGGC | 977440 | 1.7795281 | 6.4361334 | 1 |
| CTGGC | 1406630 | 1.7698448 | 6.9754877 | 1 |
| GCCGA | 1393450 | 1.7633439 | 5.25111 | 1 |
| CCGAT | 1909500 | 1.6583014 | 7.305852 | 3 |
| CAGCG | 1191705 | 1.5080452 | 5.1980014 | 1 |
| ACCGA | 1702365 | 1.4869173 | 5.2117624 | 2 |
| CTTTC | 2418885 | 1.4333986 | 6.1631293 | 1 |
| TCCTT | 2336025 | 1.3842969 | 5.044856 | 2 |
| GATCT | 2235300 | 1.3415911 | 5.8879457 | 5 |
| CCTTT | 2130320 | 1.2623988 | 5.0353713 | 3 |
| CTCGG | 992190 | 1.2483896 | 5.2732954 | 1 |
| GGGGC | 663065 | 1.2242088 | 5.7888937 | 95-97 |
| TTTCG | 1946105 | 1.1613426 | 5.2584295 | 5 |
| TTCGT | 1890715 | 1.1282885 | 5.235955 | 6 |
| CGTAC | 1184705 | 1.0288548 | 6.5380645 | 5 |
| GTCCT | 1085485 | 0.93729734 | 6.4968133 | 1 |
| GTACA | 1534990 | 0.9265741 | 5.2429233 | 6 |
| TCGTA | 1531100 | 0.9189416 | 5.1137733 | 7 |