![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-03-R2_val_2.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 17038723 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 38 |
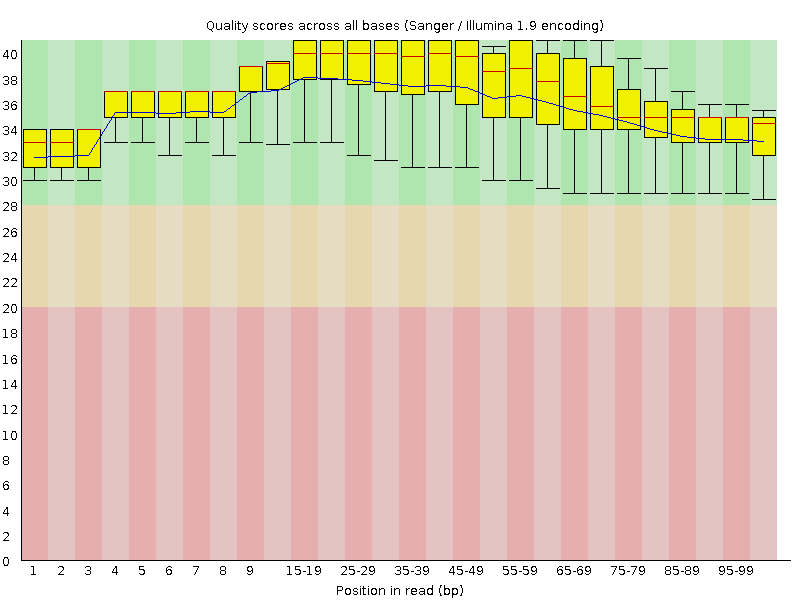
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
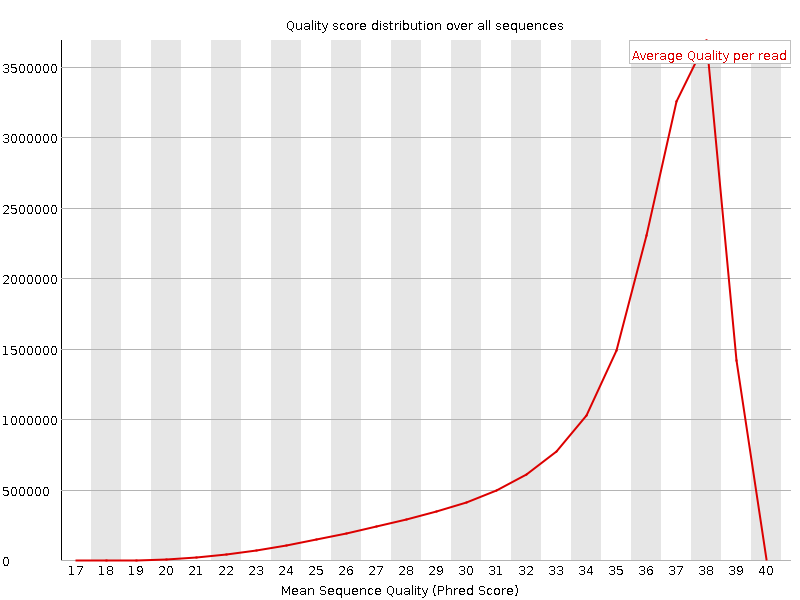
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
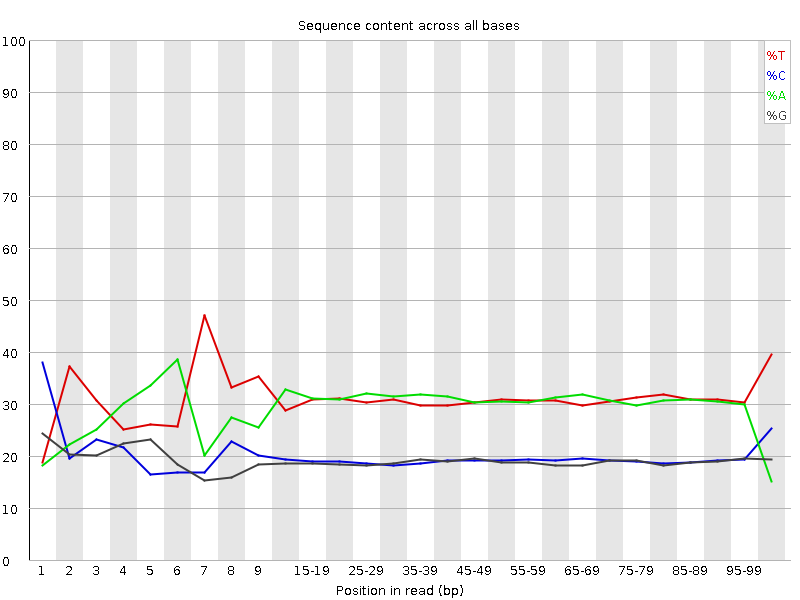
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
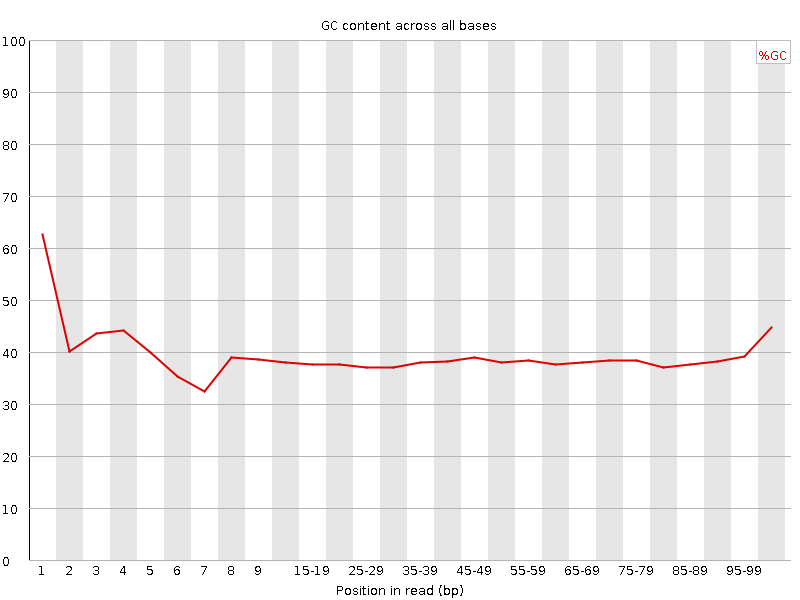
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
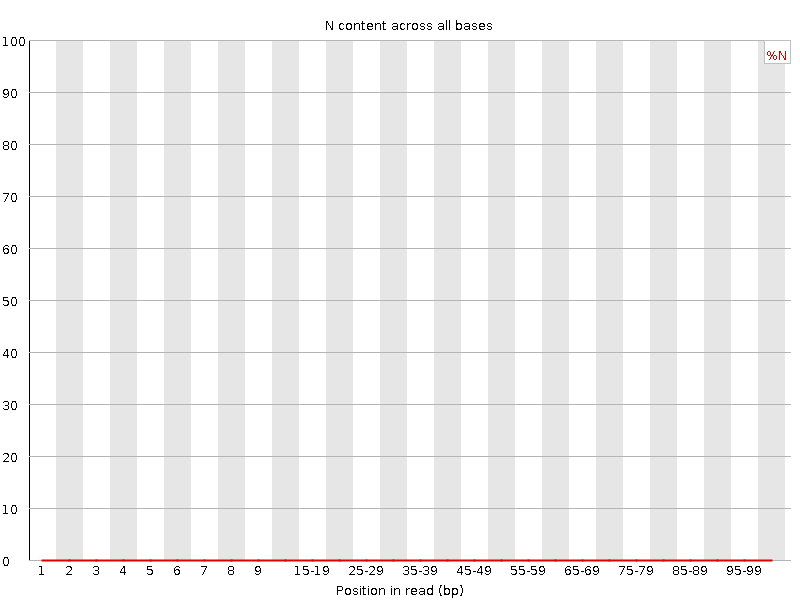
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
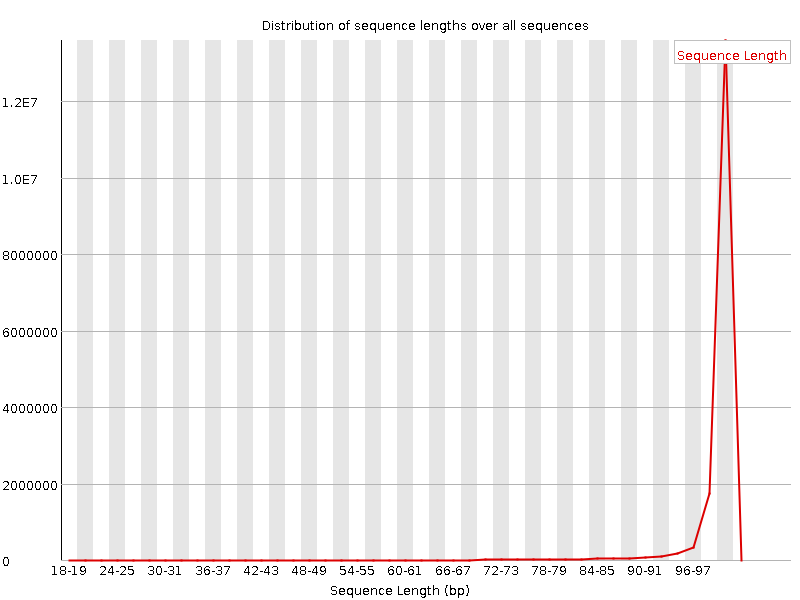
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 121845 | 0.7151064079156636 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 103441 | 0.6070936184595523 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 86073 | 0.5051610968732809 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 70426 | 0.4133290974916371 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 48255 | 0.2832078436864077 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 44989 | 0.2640397405368935 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 37761 | 0.2216187210743434 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 33433 | 0.1962177564598004 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 30354 | 0.17814715339876117 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 28946 | 0.1698836233208322 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 28841 | 0.16926737995564572 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 28299 | 0.16608639039439752 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 27248 | 0.15991808775810254 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 25115 | 0.1473995439681718 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 23523 | 0.13805612075505894 | No Hit |
| CCTCGATGTTGAATTAAAATAAAAATTAGATGTAGATGTCTAATATTTAG | 23057 | 0.1353211740105171 | No Hit |
| CAAGTTTTTAATTAAAAAACAATTGATTATGCTACCTTTGCACAGTCATT | 23050 | 0.13528009111950468 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 21726 | 0.12750955573372488 | No Hit |
| GGCAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATA | 21253 | 0.12473352609817061 | No Hit |
| CTTAAACCTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTG | 20036 | 0.117590972046438 | No Hit |
| GCTACCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGG | 19901 | 0.1167986591483411 | No Hit |
| CCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGA | 19858 | 0.11654629281783618 | No Hit |
| TTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCTT | 19725 | 0.11576571788859999 | No Hit |
| GTCTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATT | 19193 | 0.11264341817165524 | No Hit |
| CTCCCAACTAAATATAATTCATATTATTAATGATACAAAAATTTTTAATA | 18747 | 0.11002585111572036 | No Hit |
| CCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTAAA | 18160 | 0.10658075725510649 | No Hit |
| GTTGCTTTTAGCCCTTGAGATCCGTGAATTTGTTAAATCCGTCAGTTGTT | 18013 | 0.10571801654384547 | No Hit |
| GTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATG | 17887 | 0.1049785245056217 | No Hit |
| AAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTG | 17509 | 0.10276004839095043 | No Hit |
| TTATAATTAAGAAAGAATTAATTACCTTAGGGATAACAGCGTAATATTTT | 17352 | 0.1018386178353859 | No Hit |
| TAAAAACATAGCTTTTAGATTATAATTTAAGGTTATTTCTGCCCCATGAG | 17231 | 0.10112847071931388 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
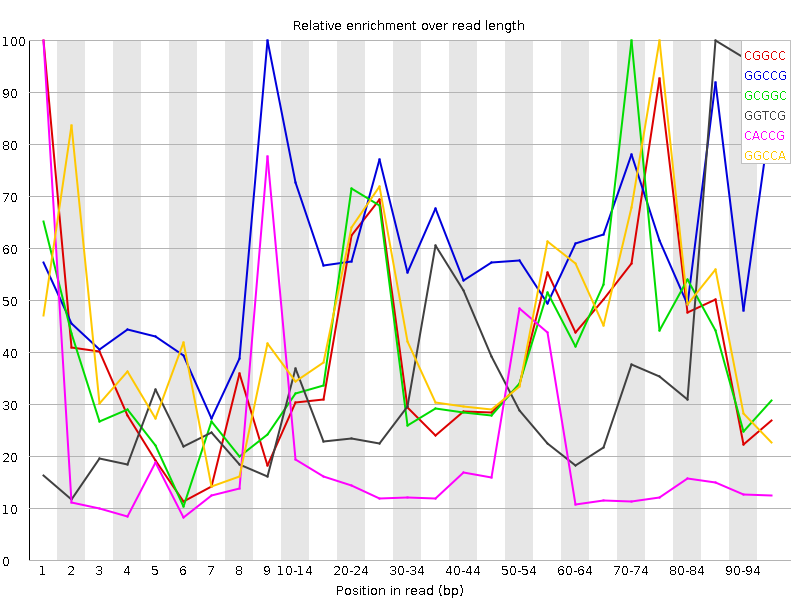
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1540980 | 3.5588174 | 8.238988 | 1 |
| GGCCG | 1431680 | 3.3858502 | 5.4986815 | 9 |
| GCGGC | 1405375 | 3.32364 | 7.6834226 | 70-74 |
| GGTCG | 1673725 | 2.5048037 | 6.389794 | 85-89 |
| CACCG | 1709605 | 2.4591293 | 13.12261 | 1 |
| GGCCA | 1659290 | 2.4441113 | 5.145349 | 75-79 |
| CGGCG | 1029385 | 2.434443 | 8.909226 | 1 |
| CGCCG | 1040110 | 2.4020824 | 8.128046 | 1 |
| CTGGC | 1629780 | 2.381801 | 10.534665 | 1 |
| CGATC | 2598655 | 2.3653858 | 12.036004 | 4 |
| CCGGC | 937205 | 2.1644282 | 7.558115 | 1 |
| CCTCG | 1500230 | 2.1410222 | 6.9759183 | 1 |
| GGCTT | 2247960 | 2.0788963 | 6.7617297 | 3 |
| CCGAT | 2259965 | 2.0570984 | 10.605834 | 3 |
| GGGGC | 815350 | 1.9745991 | 13.105465 | 95-97 |
| CTGCG | 1340325 | 1.958784 | 5.0947533 | 70-74 |
| TGGCT | 2060060 | 1.9051279 | 6.571275 | 45-49 |
| CGACG | 1283020 | 1.8898706 | 5.792522 | 1 |
| GACCT | 2068595 | 1.8829067 | 8.475262 | 95-97 |
| ACCGA | 2019770 | 1.853009 | 8.555841 | 2 |
| GCCGA | 1253380 | 1.8462114 | 5.5004687 | 1 |
| AGGTC | 1943625 | 1.8116693 | 5.413034 | 95-97 |
| CGCGG | 763115 | 1.804728 | 5.858549 | 1 |
| CCGCG | 778935 | 1.7989117 | 5.5263753 | 1 |
| GCAAA | 3051780 | 1.7857426 | 5.335418 | 2 |
| CAGCG | 1128030 | 1.6615725 | 6.9843245 | 1 |
| GTCGC | 1084985 | 1.585624 | 7.216504 | 95-97 |
| ATATT | 6938235 | 1.5751244 | 5.1770897 | 4 |
| CGCAA | 1686995 | 1.5477093 | 5.2687106 | 1 |
| CTTTC | 2757075 | 1.5386307 | 9.939292 | 1 |
| GATCT | 2610670 | 1.5037414 | 7.4953003 | 5 |
| CGAAC | 1597520 | 1.4656216 | 6.2620845 | 90-94 |
| TCCTT | 2548340 | 1.4221427 | 7.7188306 | 2 |
| CCTTT | 2535250 | 1.4148376 | 7.749255 | 3 |
| CTAAA | 3843710 | 1.3898594 | 6.0313015 | 8 |
| CAAAT | 3600435 | 1.3018928 | 5.84746 | 9 |
| AGACC | 1401140 | 1.2854556 | 7.1858554 | 95-97 |
| ACACC | 1392520 | 1.247567 | 5.504711 | 8 |
| CAGAC | 1355005 | 1.2431297 | 6.7746177 | 95-97 |
| CTCGT | 1323095 | 1.194874 | 5.098711 | 1 |
| CTCGG | 809365 | 1.1828259 | 5.7573805 | 1 |
| CTCAT | 2096160 | 1.1790506 | 6.79535 | 1 |
| TTTCG | 1955090 | 1.1172893 | 8.301423 | 2 |
| CGTAC | 1220535 | 1.1109732 | 12.3053 | 5 |
| TTCGT | 1941900 | 1.1097516 | 8.2058 | 3 |
| CTCCC | 731140 | 1.018945 | 6.031819 | 1 |
| GTCCT | 1019275 | 0.92049724 | 11.142479 | 1 |
| GGGTC | 600530 | 0.8987197 | 5.2622423 | 2 |
| TCGTA | 1557020 | 0.8968408 | 7.886001 | 7 |
| GTACA | 1531680 | 0.88922447 | 8.417034 | 6 |
| ACAAA | 2313985 | 0.8433407 | 5.463969 | 8 |
| TACAA | 2204235 | 0.7970364 | 5.8930507 | 7 |