![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-07-R2_val_2.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 17635978 |
| Filtered Sequences | 0 |
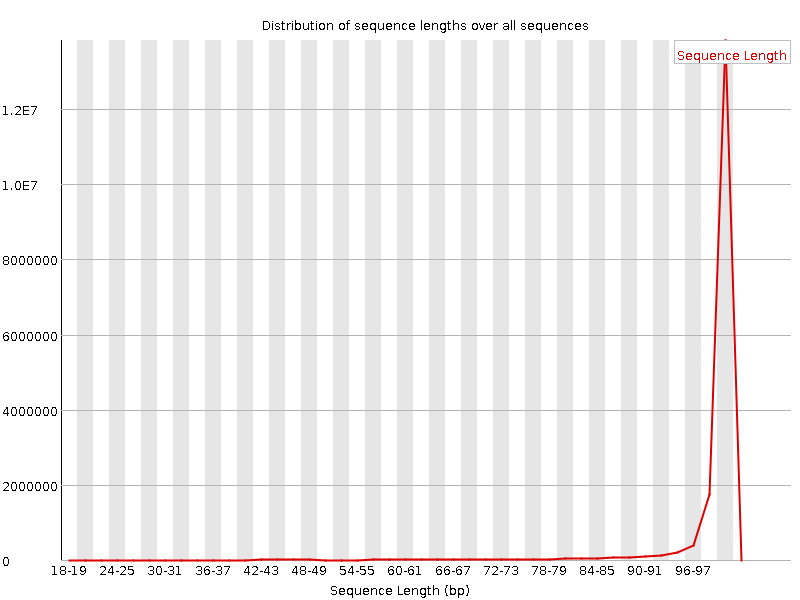
| Sequence length | 20-101 |
| %GC | 40 |
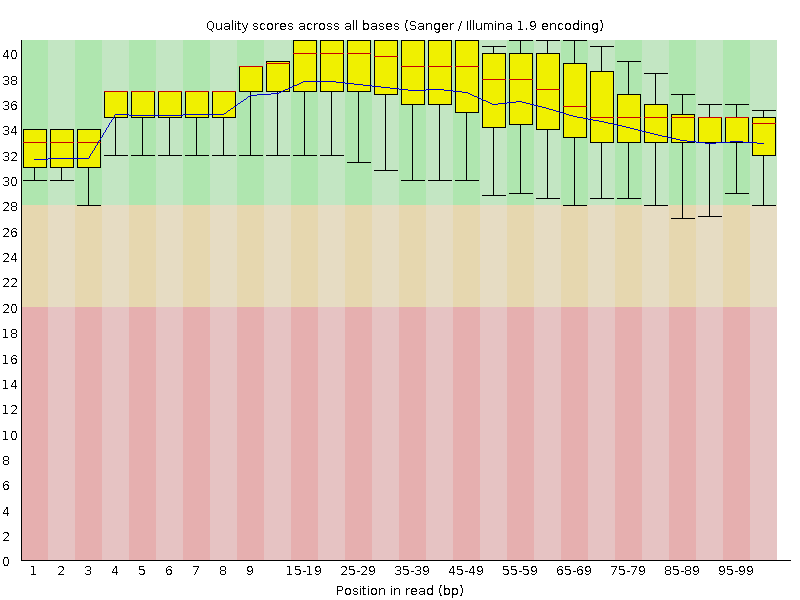
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores

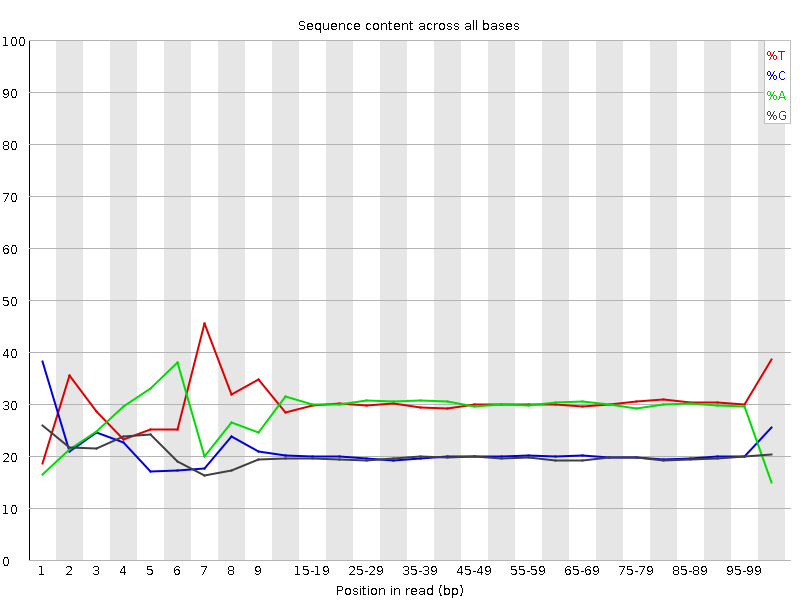
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
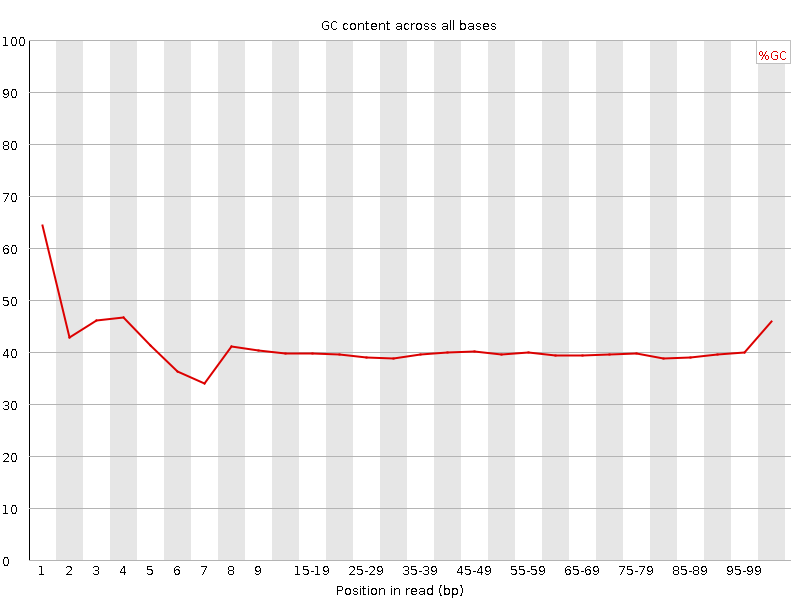
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
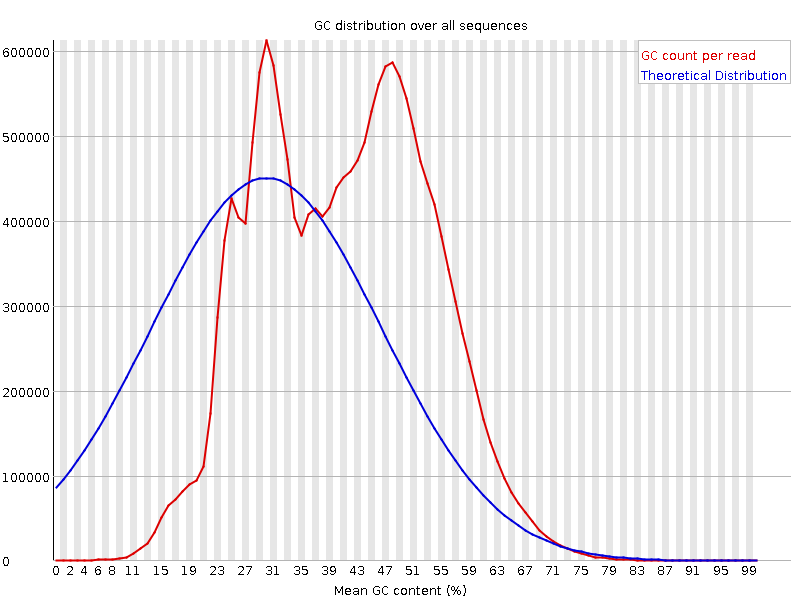
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
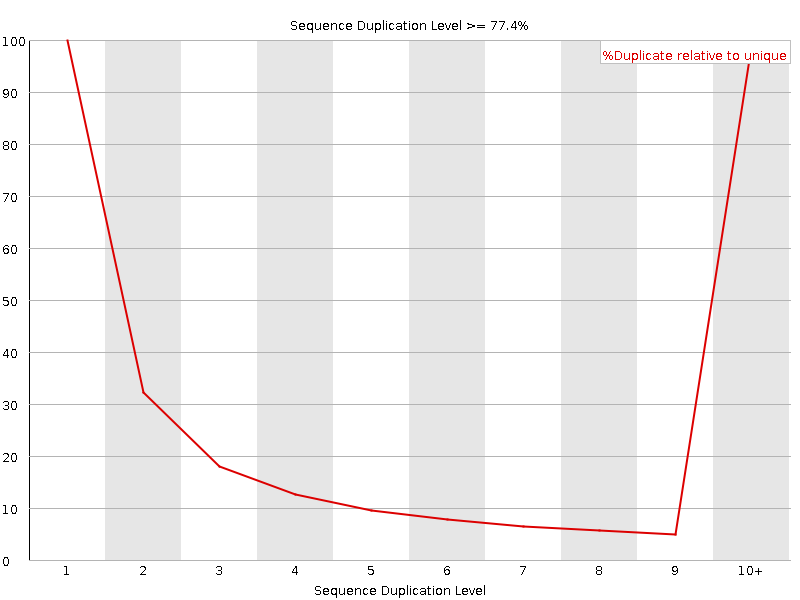
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAACACTAATTATAGATAGAAACCGATCTGGCT | 82351 | 0.46694887008818 | No Hit |
| GTCCTTTCGTACAAATAATTTAACACTAATTATAGATAGAAACCGATCTG | 74150 | 0.4204473378227167 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 62460 | 0.3541623832826283 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 38363 | 0.21752692138763155 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 30757 | 0.1743991742334902 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 28871 | 0.16370512596466158 | No Hit |
| CAAATATTGAGCTCAACTGTATATTAAAAACATAGCTTTTAGATTATAAT | 28403 | 0.16105145969222687 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 21932 | 0.12435942027144739 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 21693 | 0.12300423599984078 | No Hit |
| CTCCCAACTAAATATAATCTATACTATTAATGATACAAAAATTATTAATA | 21404 | 0.12136554037434158 | No Hit |
| GGCAAATATTGAGCTCAACTGTATATTAAAAACATAGCTTTTAGATTATA | 20879 | 0.11838867115846936 | No Hit |
| TAAAAACATAGCTTTTAGATTATAATTTAAGGTTATTTCTGCCCTATGAA | 19889 | 0.11277514635139599 | No Hit |
| CAAGTTTTTAATTAAAAAACAATTGATTATGCTACCTTTGCACAGTCATT | 19405 | 0.11003075644571568 | No Hit |
| CTTCTCGTCCCATAATACCATTTAAGTTTTTTTACTTAAAAAATAATTTA | 19088 | 0.10823329446203664 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 18080 | 0.10251770556756194 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
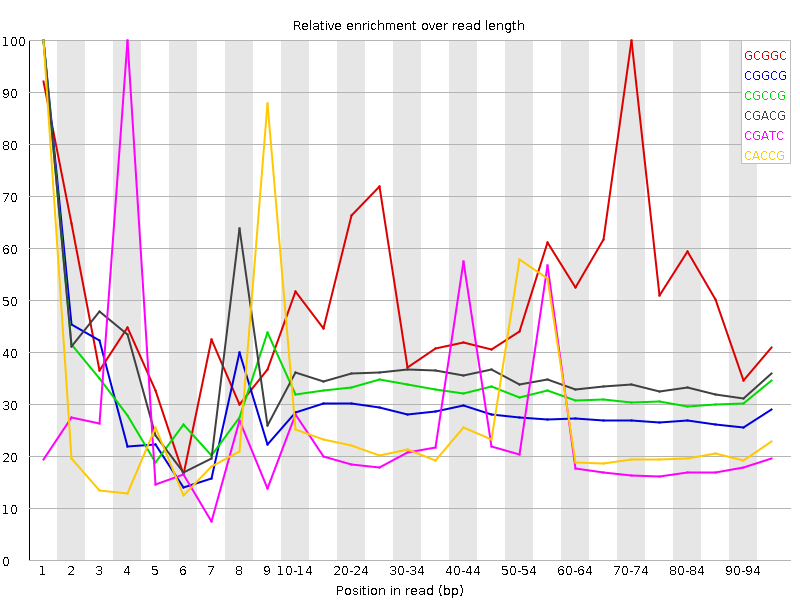
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GCGGC | 1499170 | 2.8189504 | 5.3781195 | 70-74 |
| CGGCG | 1371920 | 2.579677 | 8.951849 | 1 |
| CGCCG | 1382120 | 2.5428205 | 7.7945323 | 1 |
| CGACG | 1629910 | 2.0463912 | 5.779653 | 1 |
| CGATC | 2459315 | 2.033912 | 8.413199 | 4 |
| CACCG | 1577705 | 1.9381359 | 7.413456 | 1 |
| CGCGG | 1008530 | 1.8963803 | 6.111461 | 1 |
| CCGCG | 1012305 | 1.8624358 | 6.1606536 | 1 |
| CCTCG | 1531495 | 1.8560023 | 5.6752844 | 1 |
| CGCGA | 1401850 | 1.7600564 | 5.0216174 | 1 |
| CCGCC | 966110 | 1.7391214 | 5.013486 | 1 |
| CCGGC | 931415 | 1.7136148 | 6.28647 | 1 |
| CCGAT | 2059050 | 1.7028834 | 6.7883925 | 3 |
| CTGGC | 1354110 | 1.6771945 | 7.1143923 | 1 |
| CTTTC | 2801540 | 1.505609 | 7.452581 | 1 |
| GACCT | 1717720 | 1.4205952 | 5.6785626 | 95-97 |
| GTCGG | 1073880 | 1.3594146 | 5.4484878 | 1 |
| TCCTT | 2486115 | 1.3360928 | 5.892125 | 2 |
| GATCT | 2356775 | 1.3121867 | 5.3488564 | 5 |
| CTCGT | 1567800 | 1.2791253 | 5.1330404 | 1 |
| CCTTT | 2321570 | 1.2476628 | 5.925435 | 3 |
| CTCGG | 1006030 | 1.2460642 | 5.7063656 | 1 |
| TTTCG | 2145460 | 1.1784264 | 5.980766 | 5 |
| AGGGC | 871870 | 1.1187767 | 5.4703326 | 95-97 |
| TTCGT | 2032510 | 1.1163869 | 6.021238 | 6 |
| CGTAC | 1270570 | 1.0507915 | 7.984283 | 5 |
| GTCCT | 1177975 | 0.9610778 | 7.8294926 | 1 |
| GTACA | 1675250 | 0.9454809 | 6.149923 | 6 |
| TACAA | 2463770 | 0.9284568 | 5.0362244 | 7 |
| TCGTA | 1640000 | 0.9131063 | 6.018989 | 7 |
| CTCCC | 754295 | 0.89441174 | 6.0877666 | 1 |
| CTCAT | 1567925 | 0.85415405 | 5.179866 | 1 |