![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-17-R1_val_1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 15783637 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 42 |
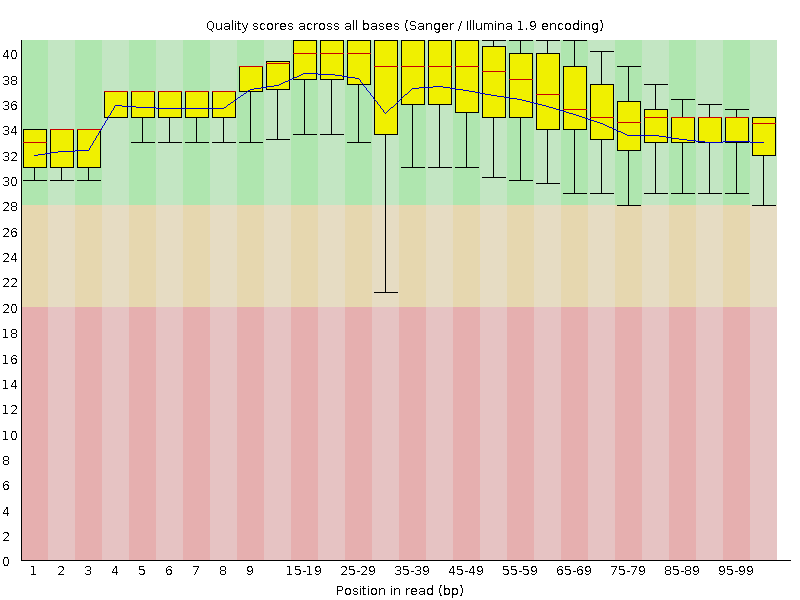
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
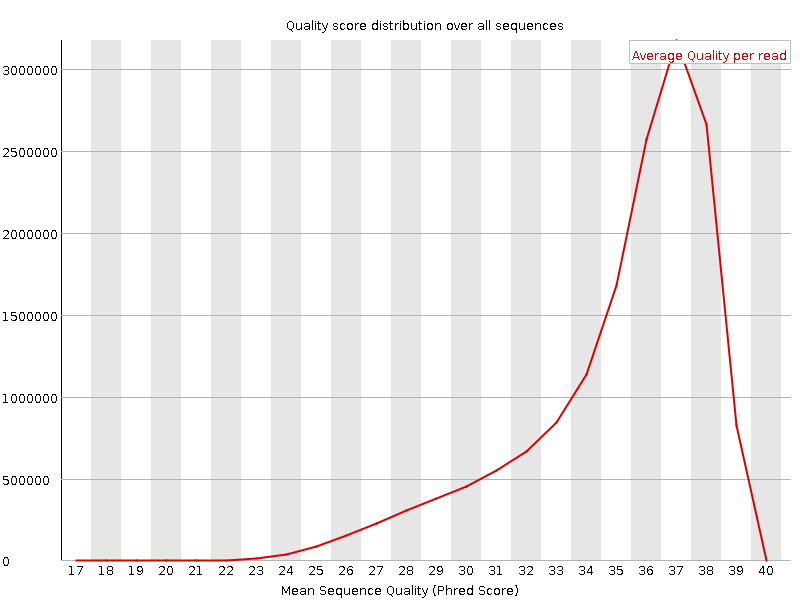
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
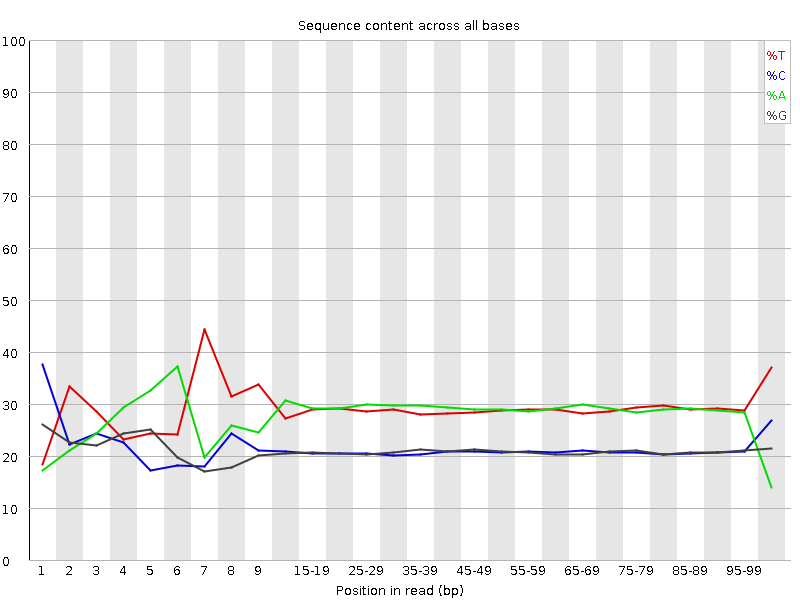
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
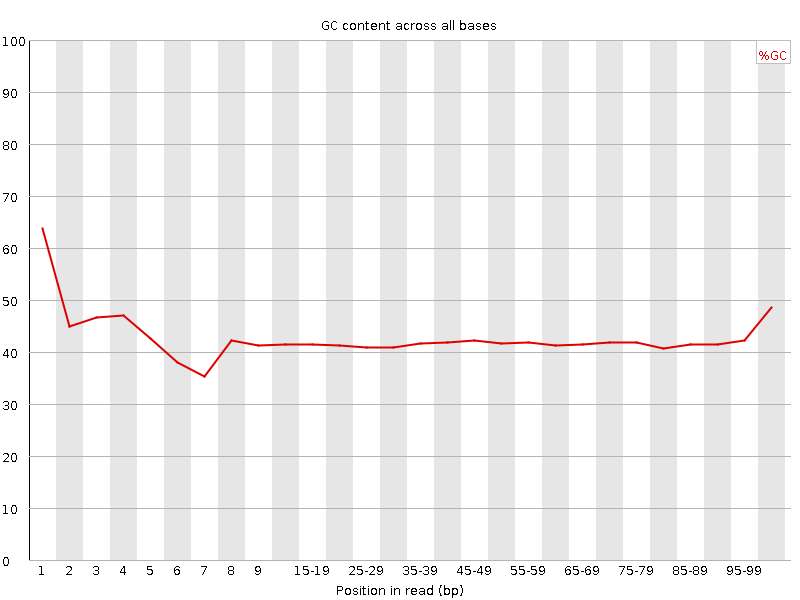
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
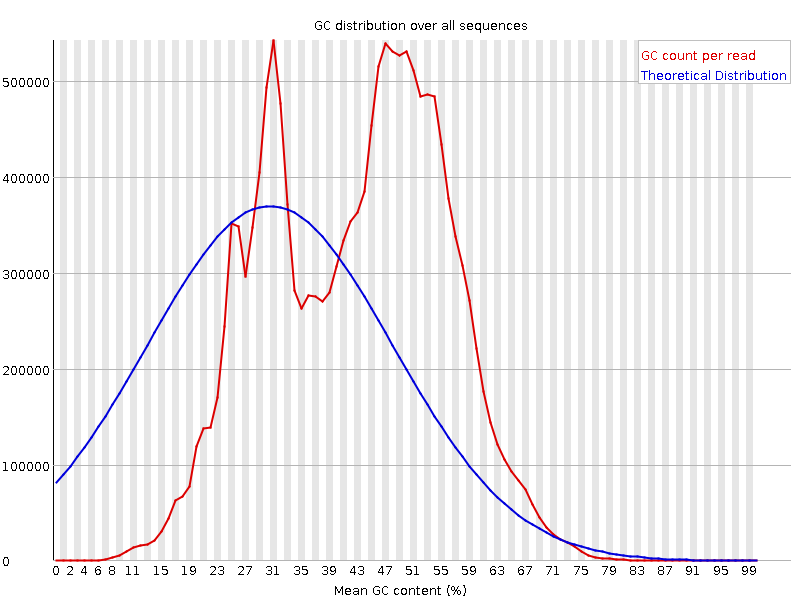
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content

![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution

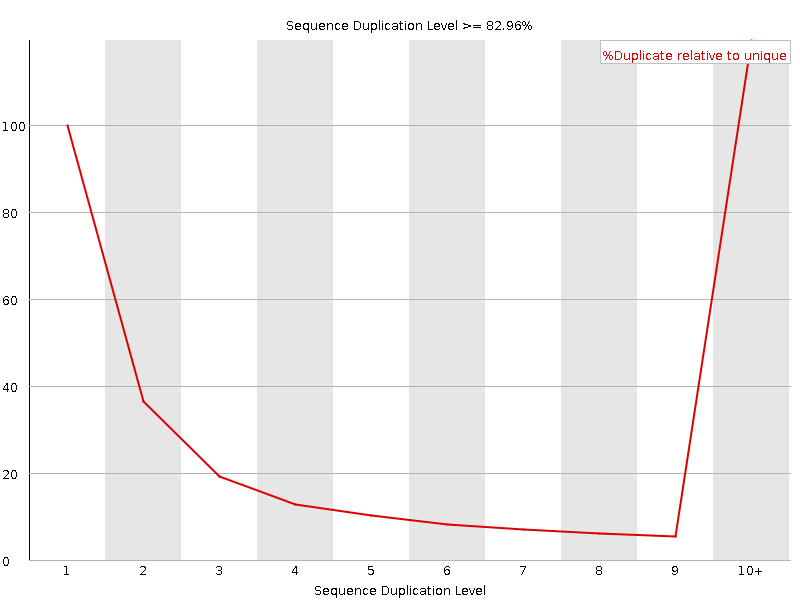
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 55260 | 0.3501094202812698 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 55135 | 0.3493174608615239 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 51712 | 0.327630444111202 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 36297 | 0.22996600846813697 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 33051 | 0.20940040625617531 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 25628 | 0.16237068807398447 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 23477 | 0.14874265037899695 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 23065 | 0.14613235213151443 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 20825 | 0.1319404393296678 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 19381 | 0.12279172411276311 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 19143 | 0.12128383337756692 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 18530 | 0.117400064383133 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 17123 | 0.10848576915447308 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 16030 | 0.10156087598821488 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
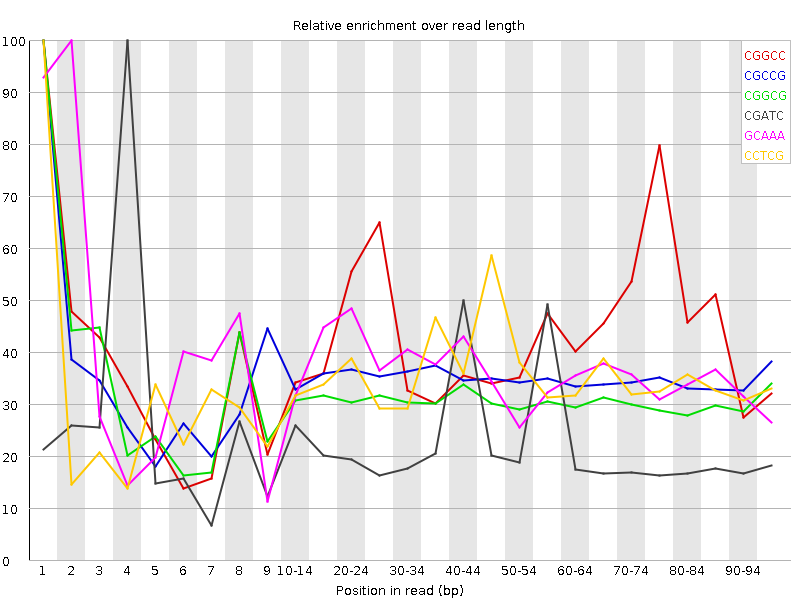
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1601955 | 2.6081722 | 6.0390806 | 1 |
| CGCCG | 1413815 | 2.3018582 | 6.561951 | 1 |
| CGGCG | 1403555 | 2.3011336 | 7.3922253 | 1 |
| CGATC | 2402720 | 2.0652292 | 9.055432 | 4 |
| GCAAA | 3129340 | 1.9757107 | 5.375399 | 2 |
| CCTCG | 1672340 | 1.9666151 | 5.5375643 | 1 |
| CGACG | 1643100 | 1.9553311 | 5.108397 | 1 |
| CACCG | 1650985 | 1.9510707 | 7.579333 | 1 |
| GCCGA | 1516530 | 1.8047096 | 5.714725 | 1 |
| CCGGC | 1085285 | 1.7669724 | 5.9263797 | 1 |
| CTGGC | 1489655 | 1.7640338 | 6.2855916 | 1 |
| ATATT | 5090080 | 1.6799798 | 5.5186915 | 4 |
| CCGAT | 1925775 | 1.6552769 | 7.489553 | 3 |
| GACCT | 1724150 | 1.4819726 | 5.31934 | 95-97 |
| ACCGA | 1714830 | 1.481226 | 5.422921 | 2 |
| TCCTT | 2387365 | 1.474883 | 5.956979 | 2 |
| CTAAA | 3223875 | 1.4701364 | 5.598322 | 8 |
| CTTTC | 2366275 | 1.461854 | 6.449536 | 1 |
| GATCT | 2251835 | 1.4077867 | 6.334753 | 5 |
| GGGGC | 829815 | 1.3699987 | 6.964967 | 95-97 |
| CCTTT | 2122935 | 1.3115216 | 5.920383 | 3 |
| TTTCG | 1851535 | 1.1518532 | 5.999513 | 5 |
| TTCGT | 1789830 | 1.1134661 | 6.024081 | 6 |
| CTCAT | 1772210 | 1.1002442 | 5.5832334 | 1 |
| CGTAC | 1165250 | 1.0015767 | 7.0531263 | 8 |
| GTCCT | 1098245 | 0.9393538 | 7.207114 | 1 |
| TCGTA | 1413790 | 0.8838636 | 5.919451 | 7 |
| GTACA | 1372685 | 0.8623953 | 5.4899254 | 6 |