![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-17-R2_val_2.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 15783637 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 41 |
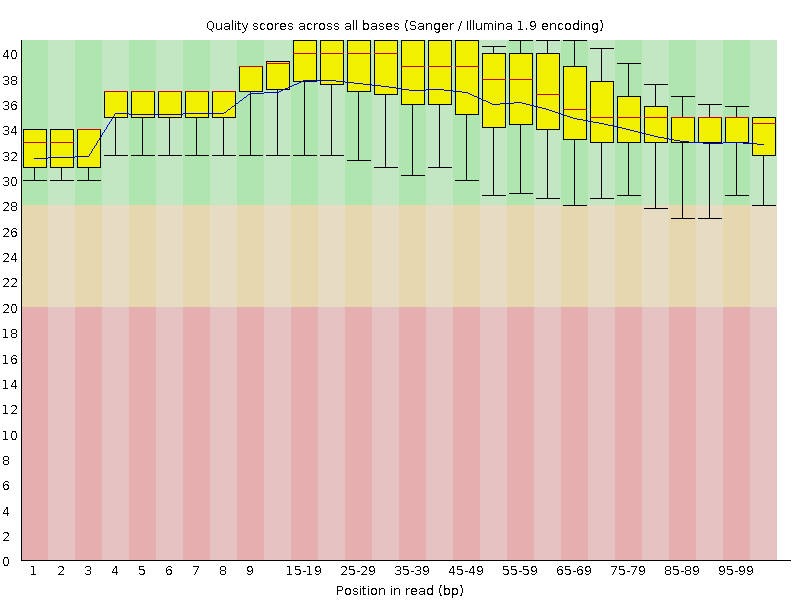
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
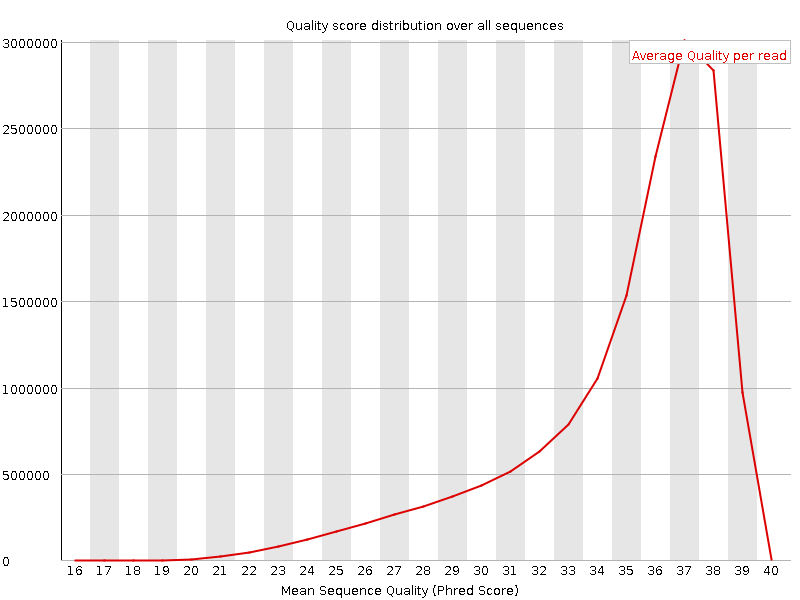
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
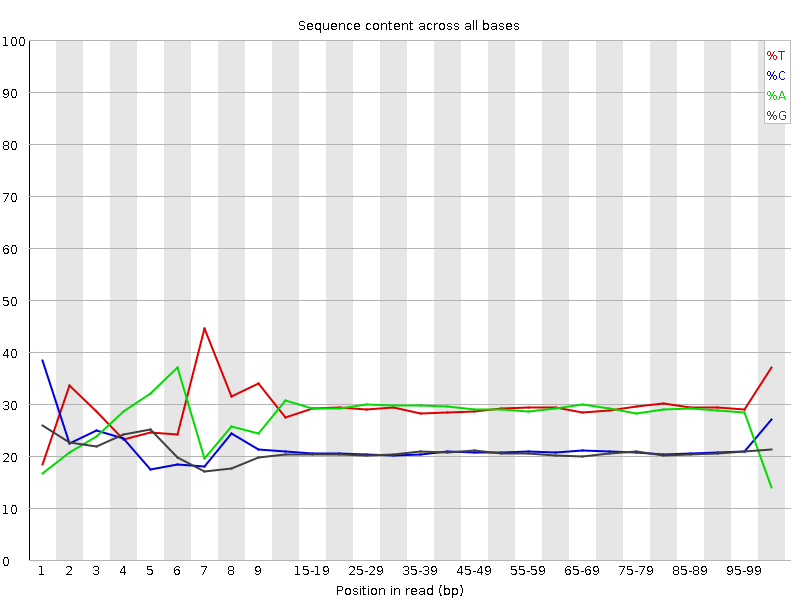
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
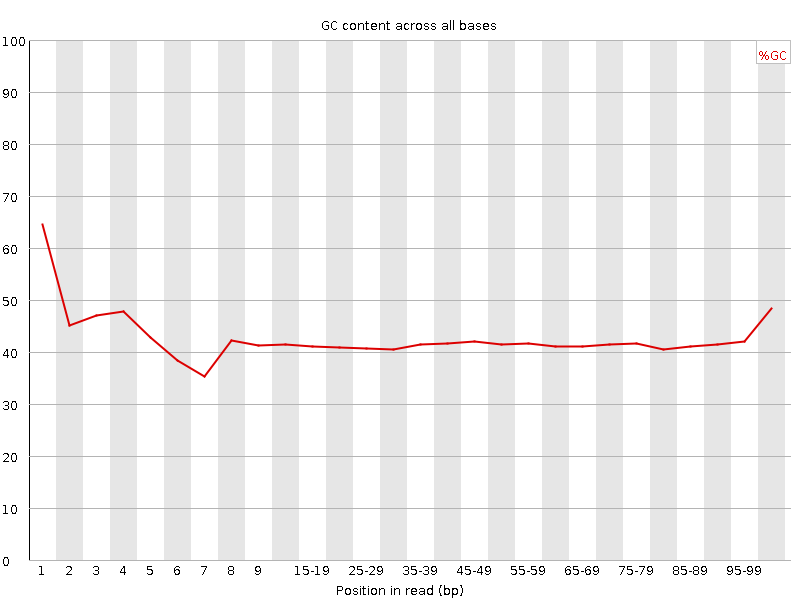
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
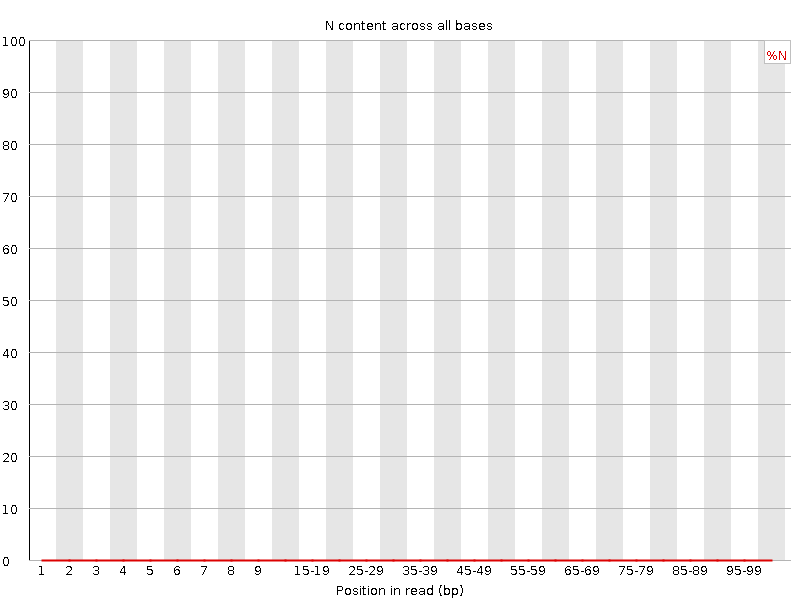
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
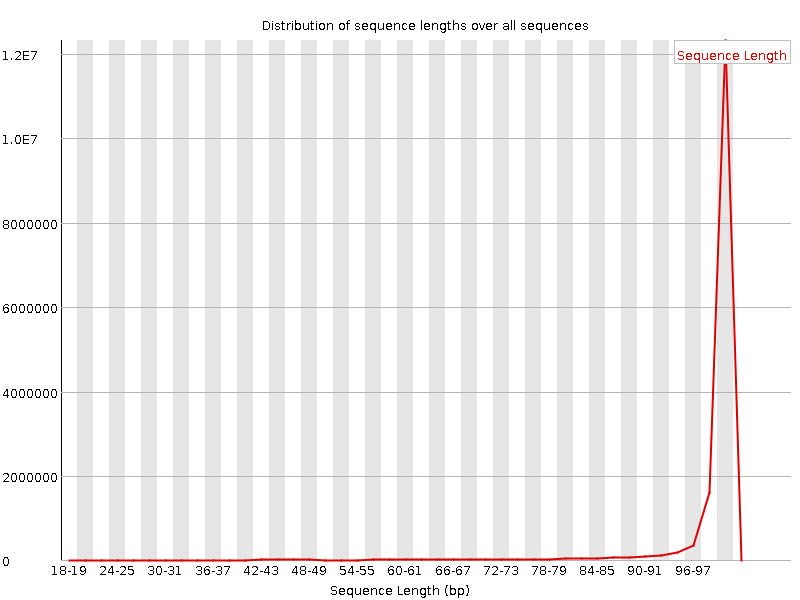
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 79667 | 0.5047442487431762 | No Hit |
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 73153 | 0.46347365946137764 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 64445 | 0.40830259844419886 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 47995 | 0.3040807388056378 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 34840 | 0.2207349294715787 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 29417 | 0.18637656200532235 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 27388 | 0.1735214767040068 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 25379 | 0.16079310490985063 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 23634 | 0.1497373514101978 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 21332 | 0.1351526267361572 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 20390 | 0.12918442054895204 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 20153 | 0.12768286548911384 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 19938 | 0.12632069528715087 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 19511 | 0.12361536190929885 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 18169 | 0.11511288557890681 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 17063 | 0.10810562863299504 | No Hit |
| GCTACCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGG | 16595 | 0.10514053256546636 | No Hit |
| CCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGA | 15950 | 0.10105402195957751 | No Hit |
| CCTCGATGTTGAATTAAAATAAAAATTAGATGTAGATGTCTAATATTTAG | 15942 | 0.10100333655671376 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
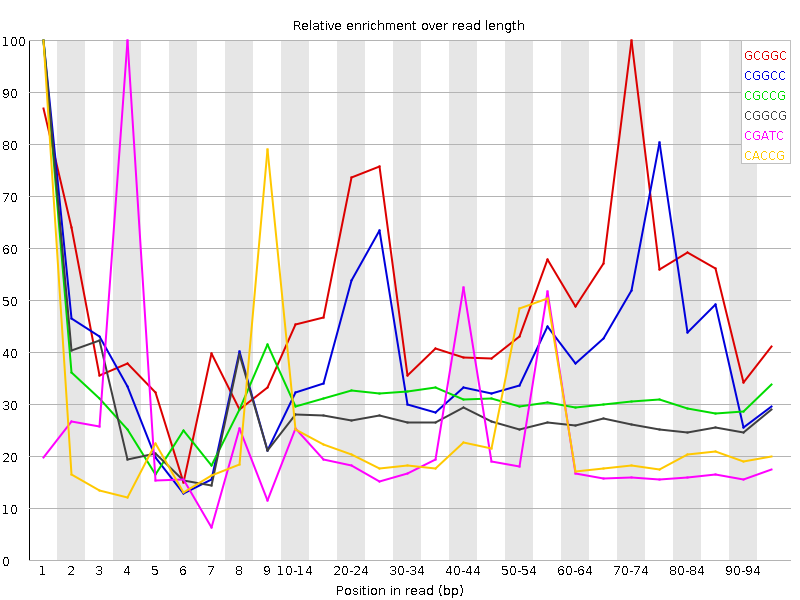
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GCGGC | 1566110 | 2.659078 | 5.104303 | 70-74 |
| CGGCC | 1595955 | 2.6553588 | 6.3987937 | 1 |
| CGCCG | 1390610 | 2.3137045 | 7.3852024 | 1 |
| CGGCG | 1357815 | 2.3054166 | 8.3751955 | 1 |
| CGATC | 2448940 | 2.1142626 | 9.499871 | 4 |
| CACCG | 1670820 | 1.9962069 | 8.2540455 | 1 |
| GCAAA | 3031655 | 1.9449779 | 5.2792444 | 2 |
| CGACG | 1588115 | 1.9362617 | 5.025532 | 1 |
| CCTCG | 1616075 | 1.9039817 | 5.9467683 | 1 |
| GCCGA | 1487625 | 1.8137423 | 5.932927 | 1 |
| CTGGC | 1497270 | 1.8001456 | 6.74024 | 1 |
| CCGGC | 1055870 | 1.7567625 | 6.067392 | 1 |
| CCGCG | 1051730 | 1.7498744 | 5.099697 | 1 |
| CGCGG | 1027485 | 1.7445536 | 5.04342 | 1 |
| CCGAT | 1989995 | 1.7180382 | 8.112718 | 3 |
| ATATT | 5107695 | 1.6534414 | 5.3715463 | 4 |
| ACCGA | 1775590 | 1.5545263 | 5.841264 | 2 |
| TCCTT | 2475390 | 1.4922738 | 6.670771 | 2 |
| CTTTC | 2467175 | 1.4873213 | 6.795196 | 1 |
| CTAAA | 3254320 | 1.4784063 | 5.8016305 | 8 |
| GATCT | 2333610 | 1.4558411 | 6.612943 | 5 |
| GACCT | 1677100 | 1.447904 | 5.6075163 | 95-97 |
| GGGGC | 832285 | 1.4420725 | 7.8620777 | 95-97 |
| CCTTT | 2192645 | 1.3218226 | 6.7007375 | 3 |
| CTCGG | 997840 | 1.1996883 | 5.1230783 | 1 |
| TTTCG | 1893725 | 1.1650054 | 6.794569 | 5 |
| TTCGT | 1838520 | 1.1310438 | 6.8249307 | 6 |
| CTCAT | 1834490 | 1.1214885 | 5.787838 | 1 |
| CGTAC | 1193255 | 1.0301821 | 8.34046 | 8 |
| GTCCT | 1124995 | 0.95776016 | 8.37416 | 1 |
| TCGTA | 1450315 | 0.9047905 | 6.7996573 | 7 |
| GTACA | 1391690 | 0.88044614 | 6.1717157 | 9 |