![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-21-R1_val_1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 14287639 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 40 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality

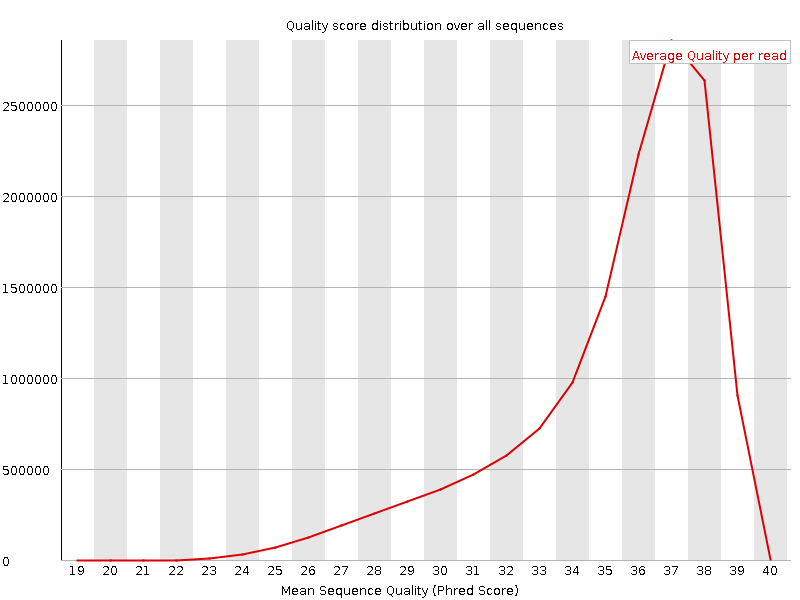
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
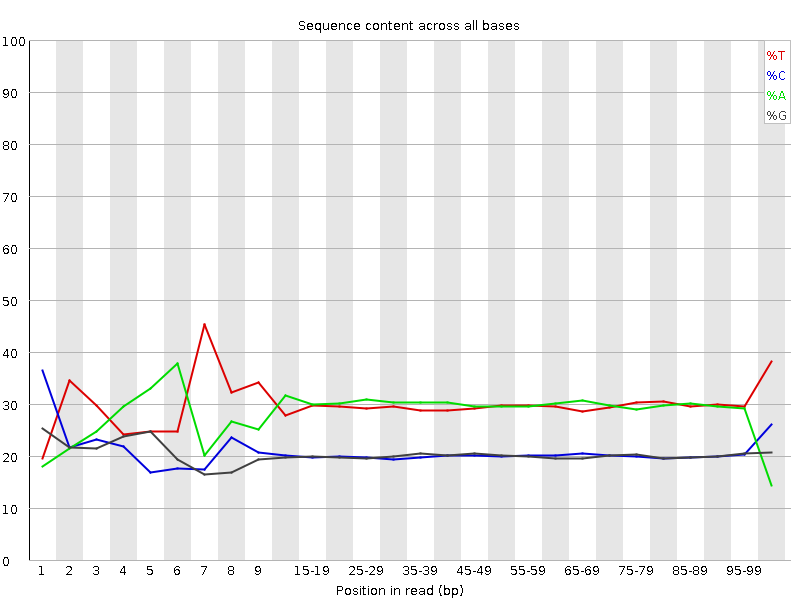
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
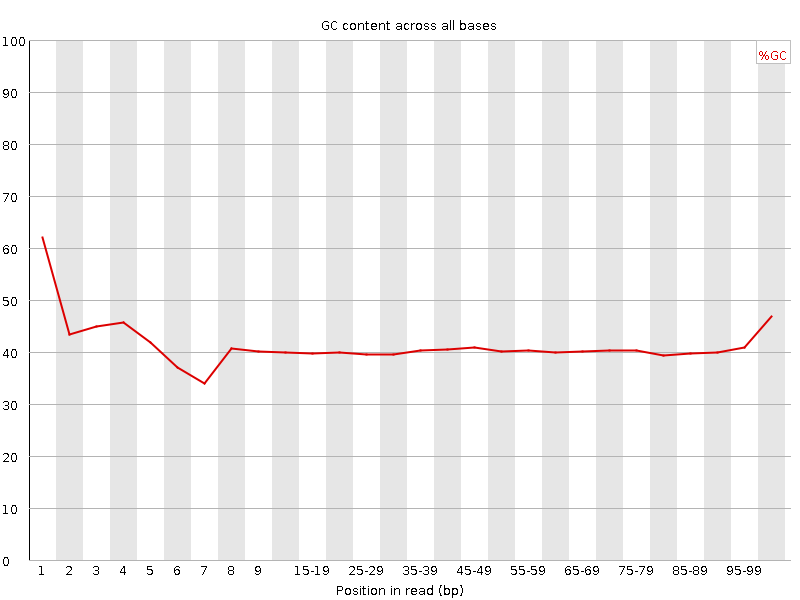
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
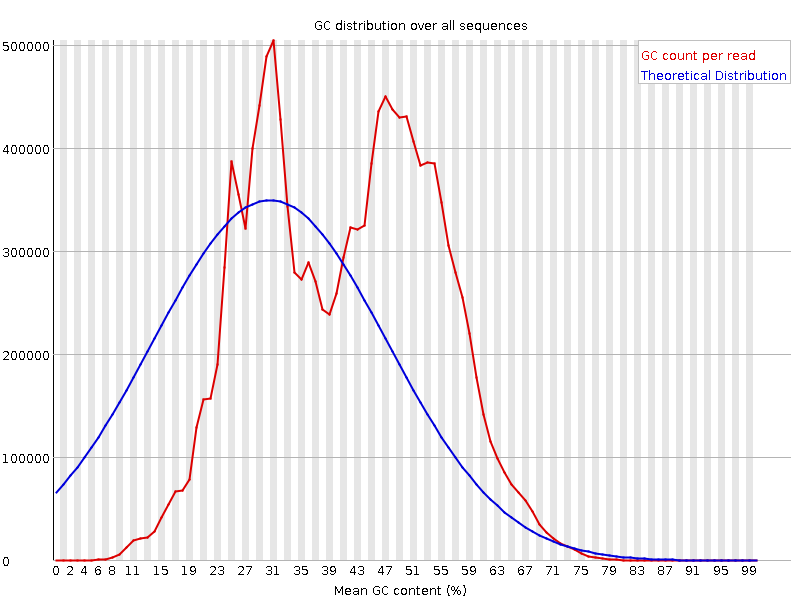
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
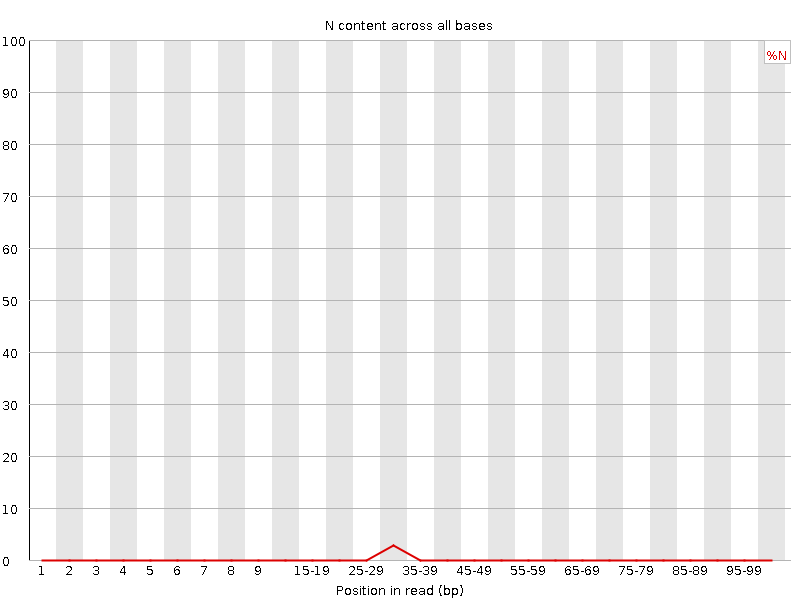
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
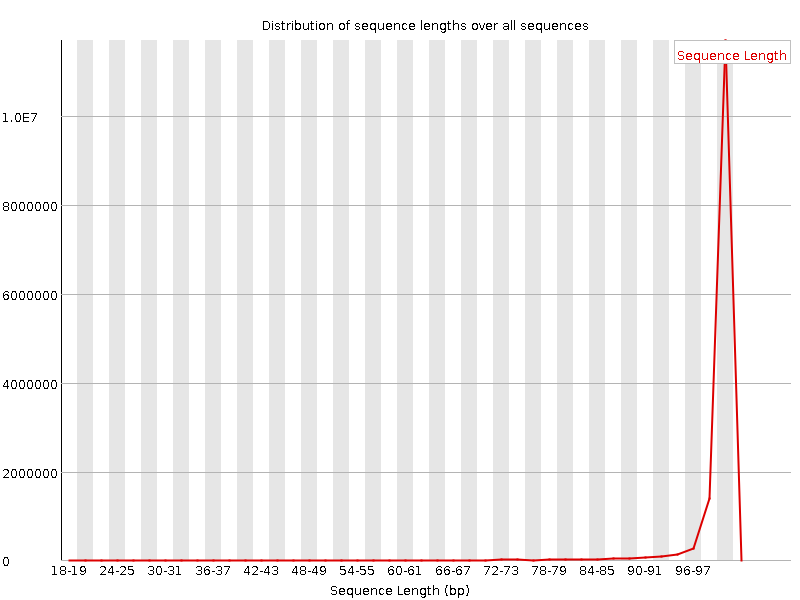
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 57676 | 0.4036776125152658 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 55393 | 0.38769876534534503 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 42622 | 0.2983138081806238 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 36071 | 0.25246298566194175 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 28580 | 0.20003304954723447 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 28497 | 0.1994521278148195 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 25676 | 0.17970778796972683 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 22819 | 0.15971148207202043 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 20113 | 0.1407720337838883 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 19050 | 0.1333320361747662 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 16680 | 0.1167442710443622 | No Hit |
| GTTGCTTTTAGCCCTTGAGATCCGTGAATTTGTTAAATCCGTCAGTTGTT | 16117 | 0.11280380194376412 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 14613 | 0.10227722019012378 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
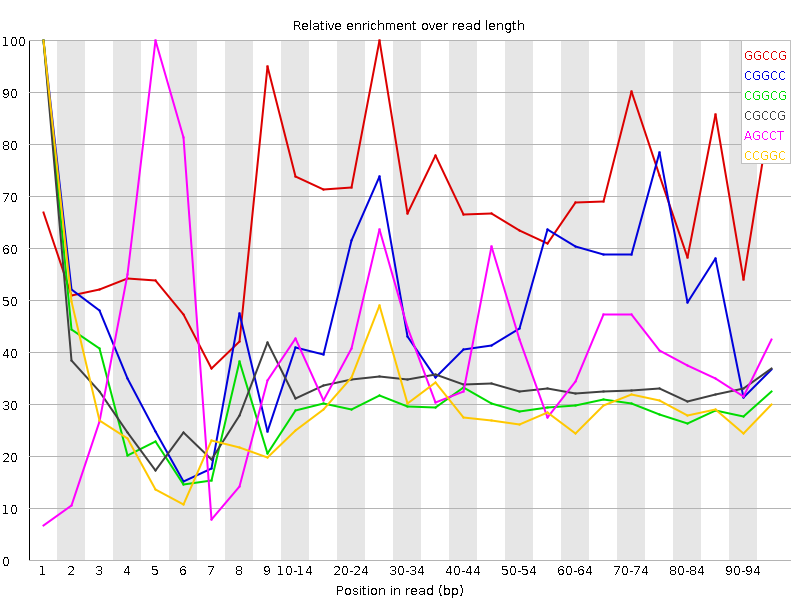
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGCCG | 1447015 | 3.1123366 | 4.4136963 | 25-29 |
| CGGCC | 1426510 | 3.0438764 | 6.0517645 | 1 |
| CGGCG | 1150900 | 2.4754329 | 8.182935 | 1 |
| CGCCG | 1145005 | 2.4432034 | 7.244816 | 1 |
| AGCCT | 2379935 | 2.3834143 | 5.911323 | 5 |
| CCGGC | 1048900 | 2.2381349 | 7.4107957 | 1 |
| GGCTT | 2168785 | 2.1799817 | 6.244444 | 3 |
| CACCG | 1482375 | 2.1629696 | 9.897942 | 1 |
| CGATC | 2130950 | 2.1340652 | 10.188101 | 4 |
| CCTCG | 1466910 | 2.1312602 | 6.058328 | 1 |
| AAGCC | 2116520 | 2.1287081 | 5.911124 | 5 |
| CTGGC | 1429125 | 2.0929773 | 8.320307 | 1 |
| GCCGA | 1317380 | 1.9376029 | 5.8452525 | 1 |
| CCGCG | 864120 | 1.8438529 | 5.092093 | 1 |
| CCGAT | 1838120 | 1.8408072 | 8.851902 | 3 |
| GCAAA | 2634100 | 1.8261122 | 5.6856008 | 2 |
| GACCT | 1788220 | 1.7908342 | 6.2928734 | 95-97 |
| CCTAA | 2608185 | 1.7861294 | 5.2808576 | 7 |
| CGCAA | 1637680 | 1.6471109 | 5.4773817 | 1 |
| CTAAA | 3342435 | 1.5777527 | 6.3858953 | 8 |
| ACCGA | 1554045 | 1.562994 | 6.907514 | 2 |
| CAGCG | 1034175 | 1.5210648 | 6.21552 | 1 |
| GGGGC | 673675 | 1.4605796 | 6.843993 | 95-97 |
| GTCGC | 992995 | 1.4542575 | 5.401988 | 95-97 |
| TCCTT | 2138515 | 1.452005 | 6.3107996 | 2 |
| GATCT | 2084015 | 1.432443 | 6.826272 | 5 |
| GCCTA | 1375335 | 1.3773457 | 5.1359982 | 6 |
| CTTTC | 2027935 | 1.3769237 | 7.1115327 | 1 |
| CCTTT | 1889920 | 1.2832144 | 6.066024 | 3 |
| AGACC | 1229890 | 1.2369726 | 5.048203 | 95-97 |
| TTCGT | 1630590 | 1.1159941 | 6.656026 | 6 |
| TTTCG | 1550770 | 1.0613644 | 6.4537425 | 5 |
| CGTAC | 1028360 | 1.0298634 | 8.133136 | 8 |
| TCGTA | 1269310 | 0.8724574 | 6.413823 | 7 |
| GTCCT | 872665 | 0.8702071 | 8.105121 | 1 |
| GTACA | 1133885 | 0.7827171 | 5.8932734 | 6 |