![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-21-R2_val_2.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 14287639 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 40 |
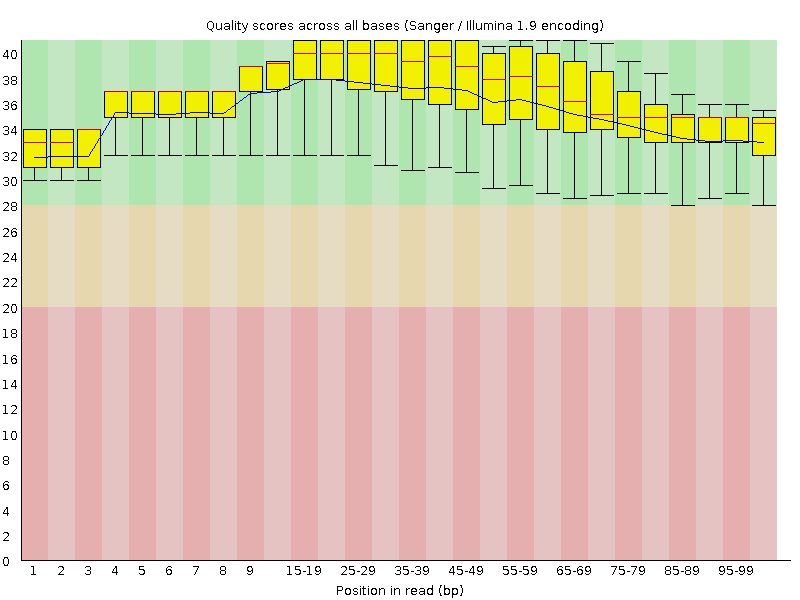
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
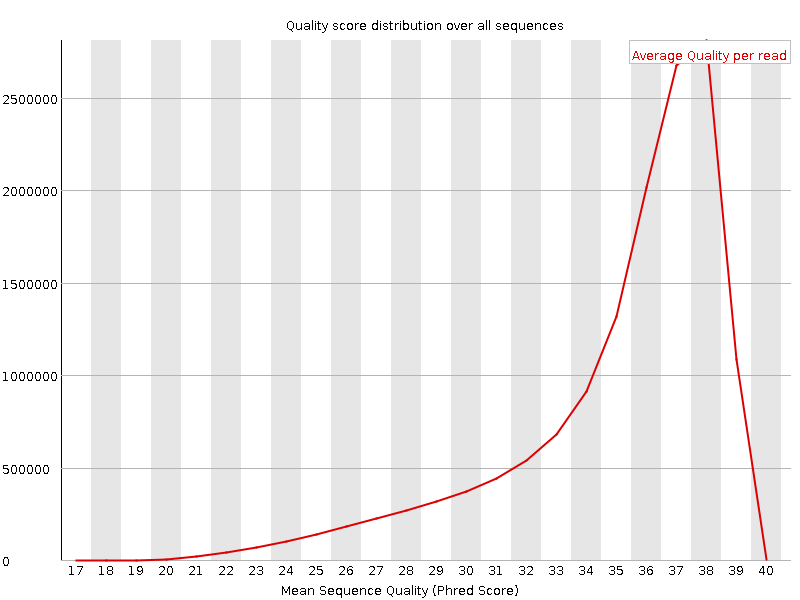
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
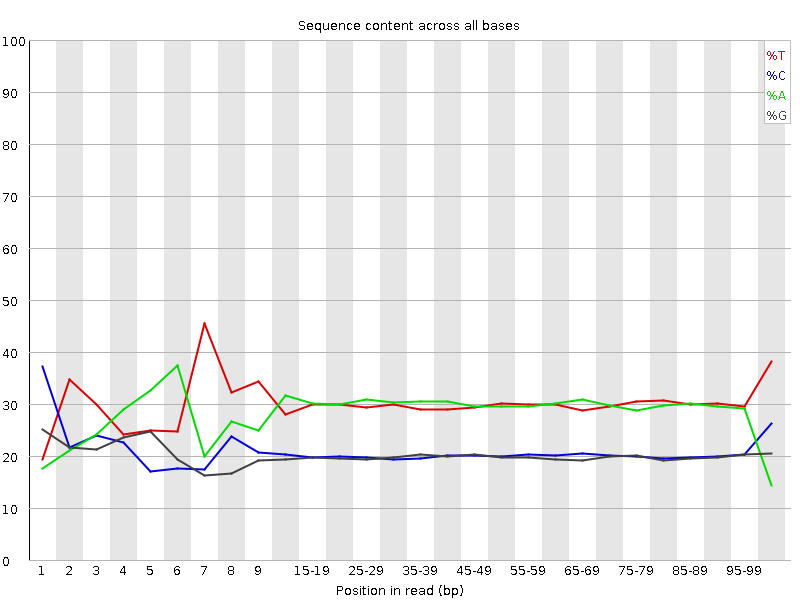
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
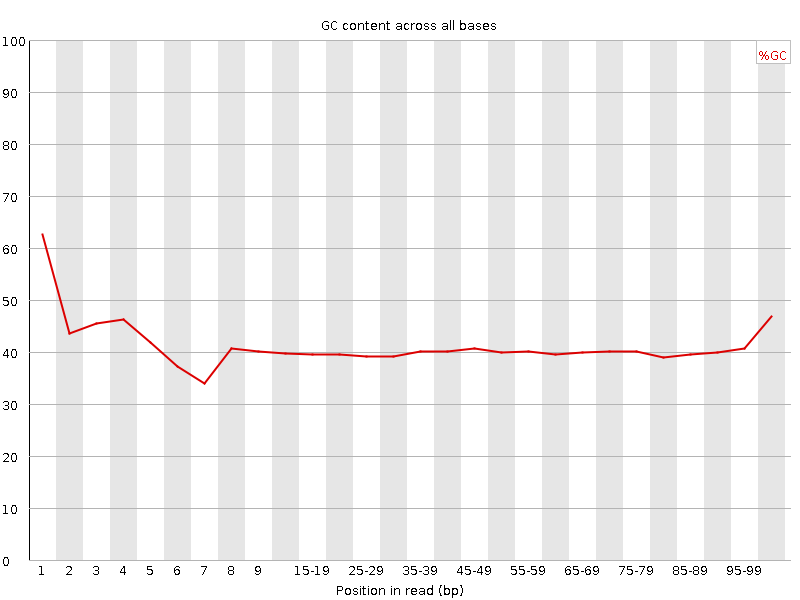
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
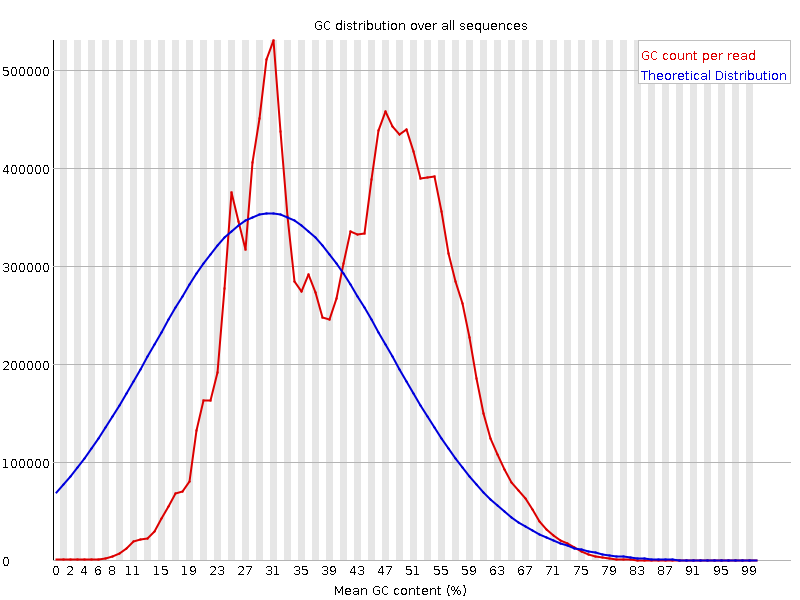
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution

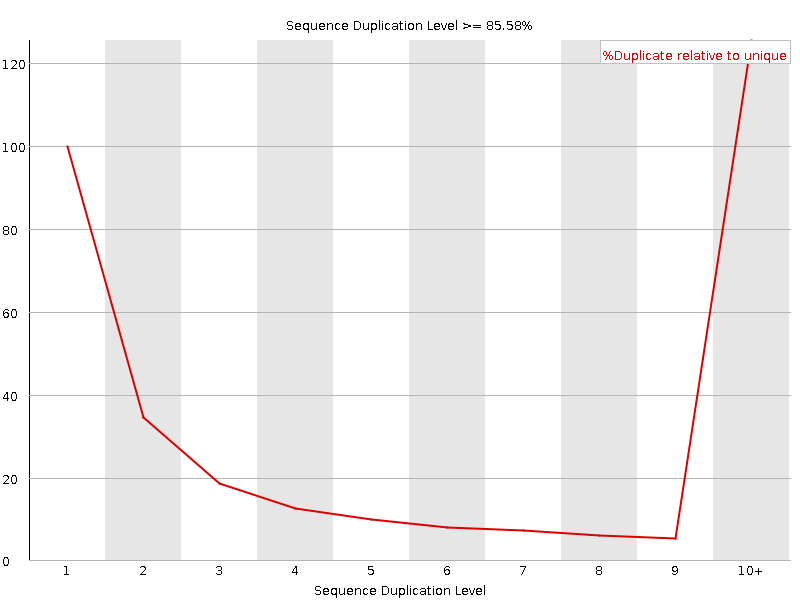
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 83135 | 0.5818666051122933 | No Hit |
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 78766 | 0.5512877250048102 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 57815 | 0.40465048144063553 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 45000 | 0.3149575657671642 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 35249 | 0.24670976079392823 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 29102 | 0.2036865573101336 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 26902 | 0.18828863187262782 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 25017 | 0.17509540939549215 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 20029 | 0.14018411299445624 | No Hit |
| GTTGCTTTTAGCCCTTGAGATCCGTGAATTTGTTAAATCCGTCAGTTGTT | 19762 | 0.13831536477090442 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 19336 | 0.13533376648164192 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 18873 | 0.13209320308274866 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 16293 | 0.11403563597876458 | No Hit |
| CCTCGATGTTGAATTAAAATAAAAATTAGATGTAGATGTCTAATATTTAG | 16201 | 0.11339172273319616 | No Hit |
| CCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTAAA | 14816 | 0.10369802876458456 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 14367 | 0.10055545216392996 | No Hit |
| CGCAAAGCCTAAATTGGCTGATGCAAAGCCTAAATTGGCTGACGCAAAGC | 14293 | 0.10003752194466839 | No Hit |
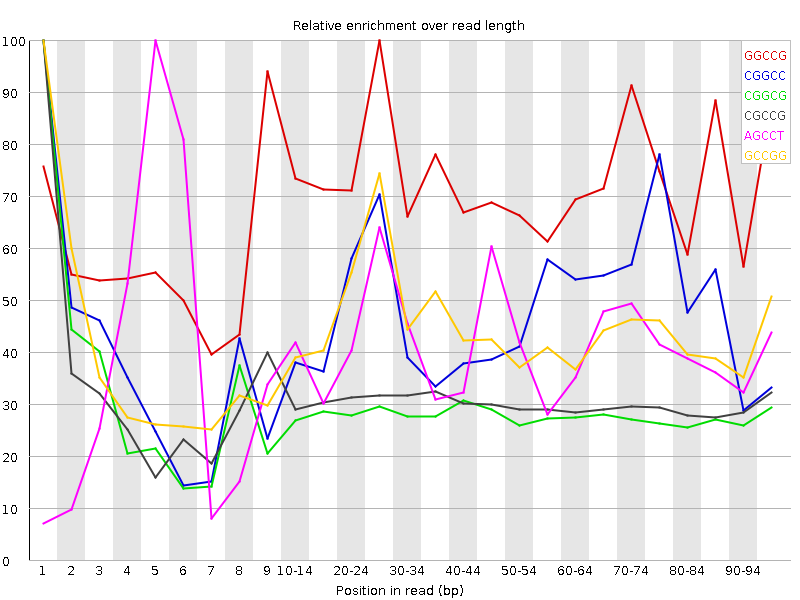
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGCCG | 1415125 | 3.1578689 | 4.4022326 | 25-29 |
| CGGCC | 1401975 | 3.0592272 | 6.4434967 | 1 |
| CGGCG | 1104565 | 2.4648502 | 8.669671 | 1 |
| CGCCG | 1122130 | 2.448582 | 8.039263 | 1 |
| AGCCT | 2313870 | 2.3273723 | 5.7013955 | 5 |
| GCCGG | 1022155 | 2.2809513 | 5.1429367 | 1 |
| GGCTT | 2207100 | 2.2443726 | 6.3057523 | 3 |
| GGTCG | 1465260 | 2.232105 | 5.701322 | 95-97 |
| CACCG | 1514510 | 2.2315075 | 11.239926 | 1 |
| CGATC | 2215200 | 2.2281265 | 11.203712 | 4 |
| CCGGC | 1020315 | 2.226413 | 7.4513493 | 1 |
| CTGGC | 1455785 | 2.1685503 | 8.641438 | 1 |
| AAGCC | 2040620 | 2.0762079 | 5.6653905 | 5 |
| CCTCG | 1397610 | 2.0357778 | 6.186933 | 1 |
| TGGCT | 1949130 | 1.982046 | 5.7007446 | 45-49 |
| CCGAT | 1949295 | 1.9606698 | 9.866384 | 3 |
| GCCGA | 1298110 | 1.9559845 | 5.8781095 | 1 |
| CGACG | 1236465 | 1.8630984 | 5.03646 | 1 |
| CCGCG | 842470 | 1.8383403 | 5.40082 | 1 |
| CGCGG | 820230 | 1.8303531 | 5.374421 | 1 |
| GCAAA | 2548525 | 1.7905236 | 5.5850415 | 2 |
| CCTAA | 2604970 | 1.7692301 | 5.094829 | 7 |
| GACCT | 1741775 | 1.7519389 | 6.7895007 | 95-97 |
| AGGTC | 1661255 | 1.7087989 | 5.035268 | 95-97 |
| ACCGA | 1645505 | 1.6742021 | 7.824359 | 2 |
| CGCAA | 1593570 | 1.6213613 | 5.3554955 | 1 |
| CTAAA | 3392195 | 1.5909075 | 6.737366 | 8 |
| GGGGC | 667540 | 1.5233663 | 7.684139 | 95-97 |
| GATCT | 2185335 | 1.5005331 | 7.4287825 | 5 |
| GTCGC | 1004260 | 1.4959545 | 6.4034653 | 95-97 |
| TCCTT | 2238255 | 1.4856882 | 7.5678606 | 2 |
| CAGCG | 984500 | 1.483439 | 5.65532 | 1 |
| CTTTC | 2122950 | 1.409152 | 7.6616926 | 1 |
| CGAAC | 1375155 | 1.3991373 | 5.3830347 | 90-94 |
| CAAAT | 2795675 | 1.3111451 | 5.122001 | 9 |
| CCTTT | 1944665 | 1.2908117 | 7.3323026 | 3 |
| AGACC | 1257940 | 1.2798781 | 5.8544984 | 95-97 |
| CTCGG | 789545 | 1.176113 | 5.031457 | 1 |
| TTCGT | 1708195 | 1.1595336 | 7.9884715 | 6 |
| CAGAC | 1135295 | 1.1550943 | 5.3758826 | 95-97 |
| TTTCG | 1619755 | 1.0994998 | 7.761055 | 5 |
| CGTAC | 1067410 | 1.0736388 | 10.2454195 | 8 |
| TCGTA | 1320870 | 0.906959 | 7.7733006 | 7 |
| GTCCT | 897595 | 0.89253575 | 9.987317 | 1 |
| GTACA | 1171365 | 0.81358266 | 7.048585 | 9 |
| TACAA | 1690815 | 0.79297626 | 5.1483407 | 7 |