![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-24-R1_val_1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 17868629 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 38 |
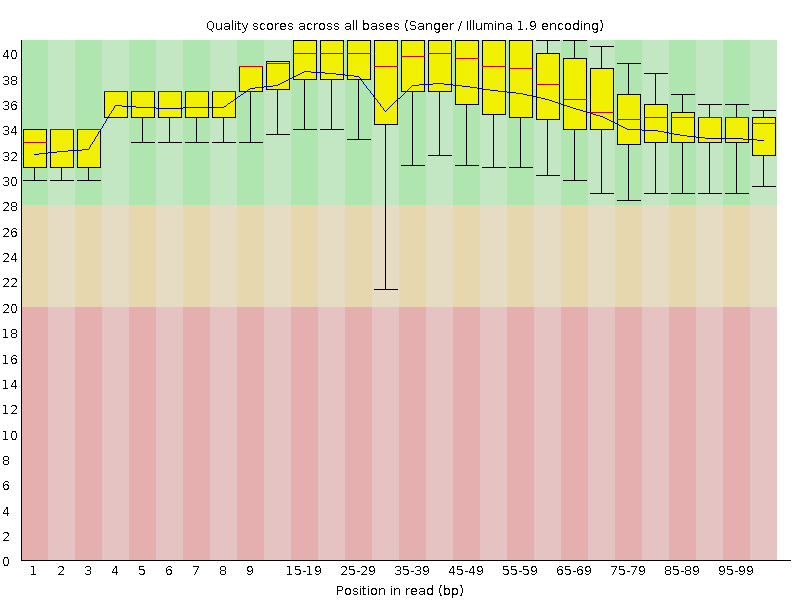
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
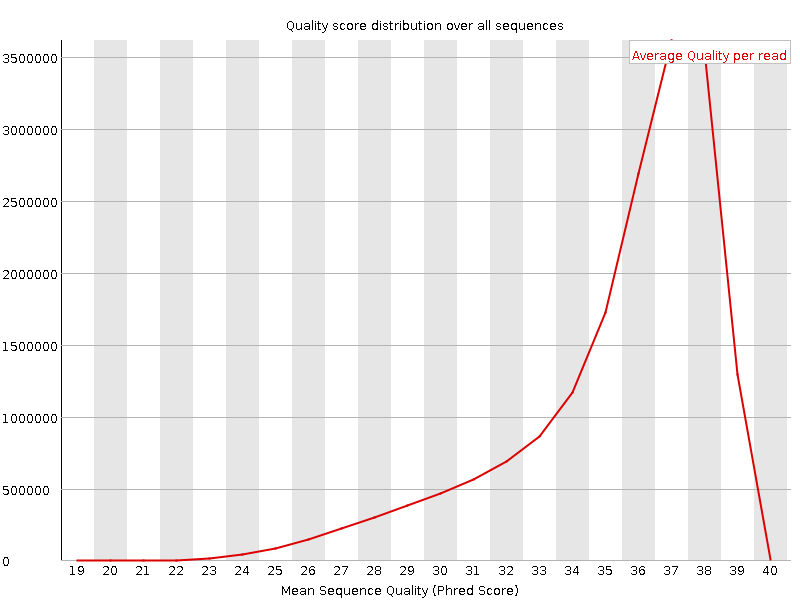
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
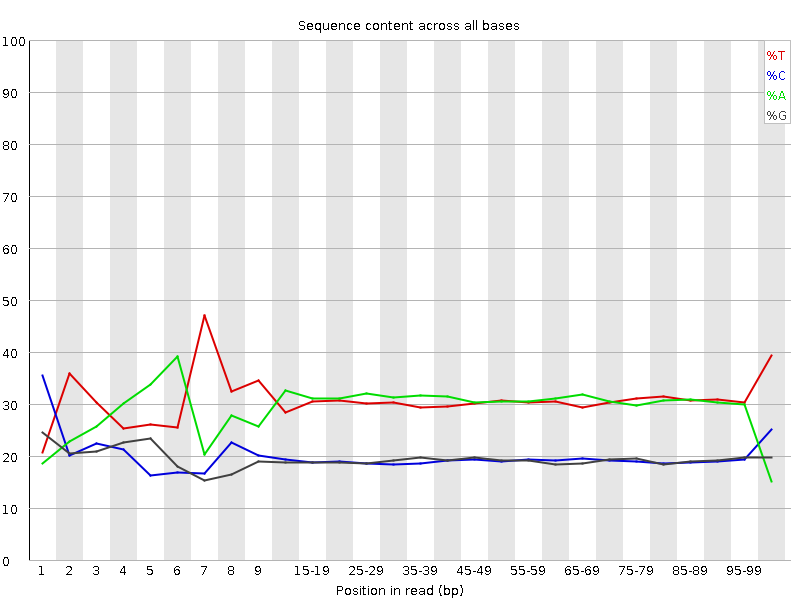
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
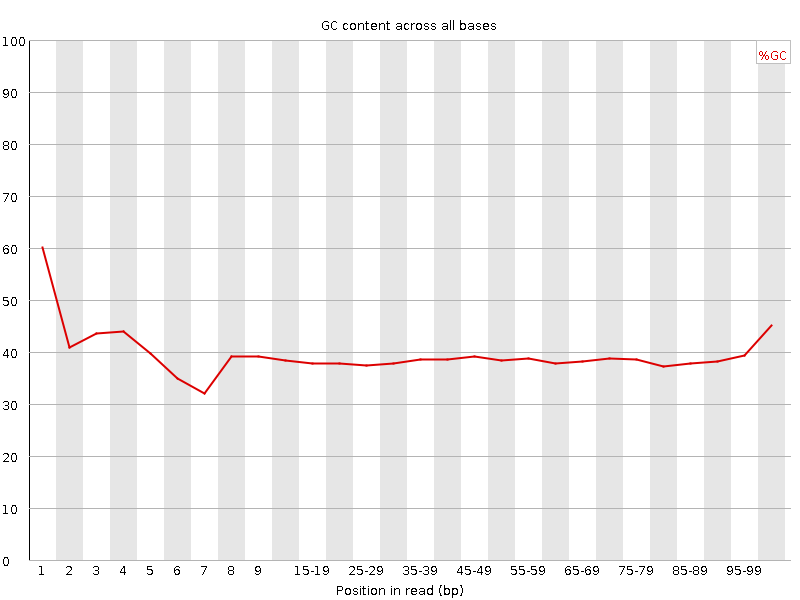
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
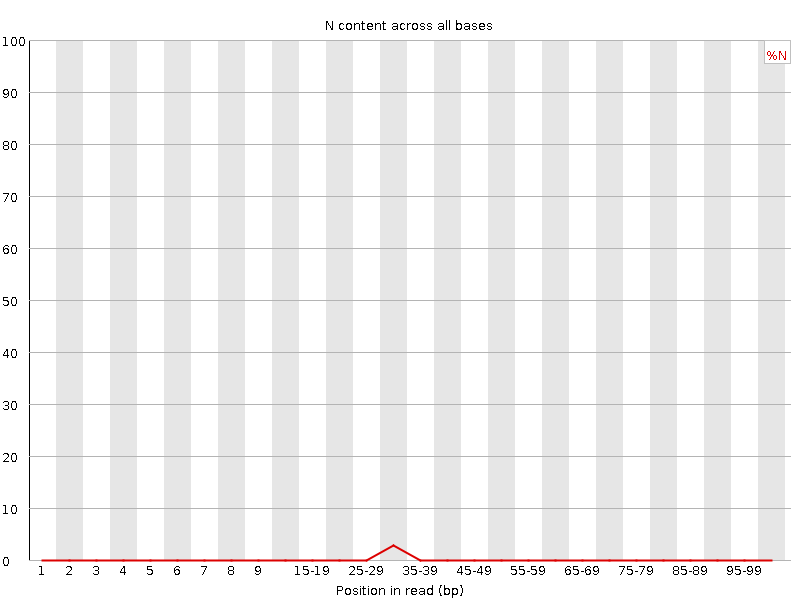
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
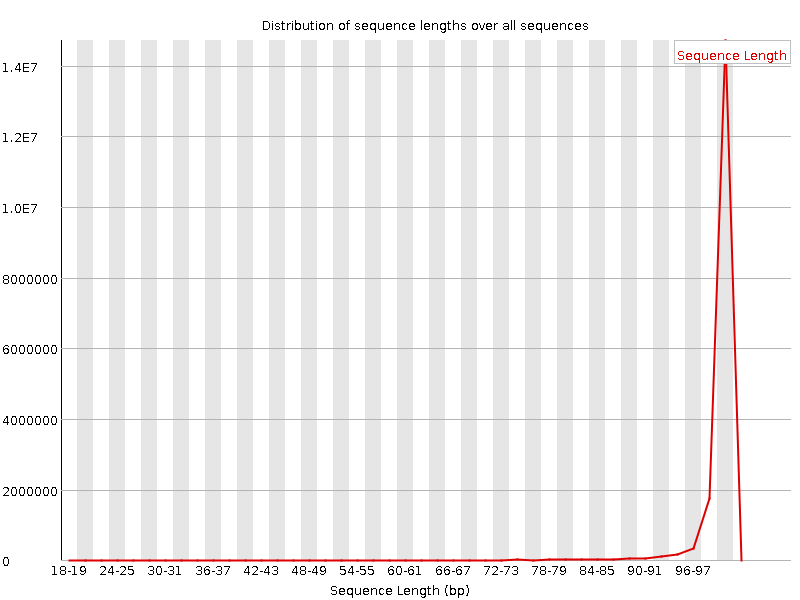
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 92065 | 0.5152325900325089 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 80698 | 0.4516183082652844 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 65663 | 0.3674764303405706 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 57508 | 0.3218377862118017 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 44652 | 0.2498904644558908 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 43992 | 0.24619684028360542 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 36009 | 0.2015207769997351 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 31378 | 0.17560384739086585 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 30034 | 0.16808228544003012 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 27839 | 0.15579818686705063 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 25689 | 0.1437659263058179 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 22093 | 0.12364127096712345 | No Hit |
| TTATAATTAAGAAAGAATTAATTACCTTAGGGATAACAGCGTAATATTTT | 21472 | 0.12016590640501854 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 20553 | 0.11502281456512416 | No Hit |
| TTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCTT | 20482 | 0.11462547014659043 | No Hit |
| CCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTAAA | 18901 | 0.10577756133388858 | No Hit |
| CCTCGATGTTGAATTAAAATAAAAATTAGATGTAGATGTCTAATATTTAG | 18741 | 0.10488213729212242 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 18383 | 0.10287862599867063 | No Hit |
| GTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATG | 18323 | 0.10254284198300831 | No Hit |
| CAAGTTTTTAATTAAAAAACAATTGATTATGCTACCTTTGCACAGTCATT | 18275 | 0.10227421477047847 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 18146 | 0.10155227913680451 | No Hit |
| AATTAATTACCTTAGGGATAACAGCGTAATATTTTTAGAAAGATCATATT | 18093 | 0.10125566992296947 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 18080 | 0.10118291671957598 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
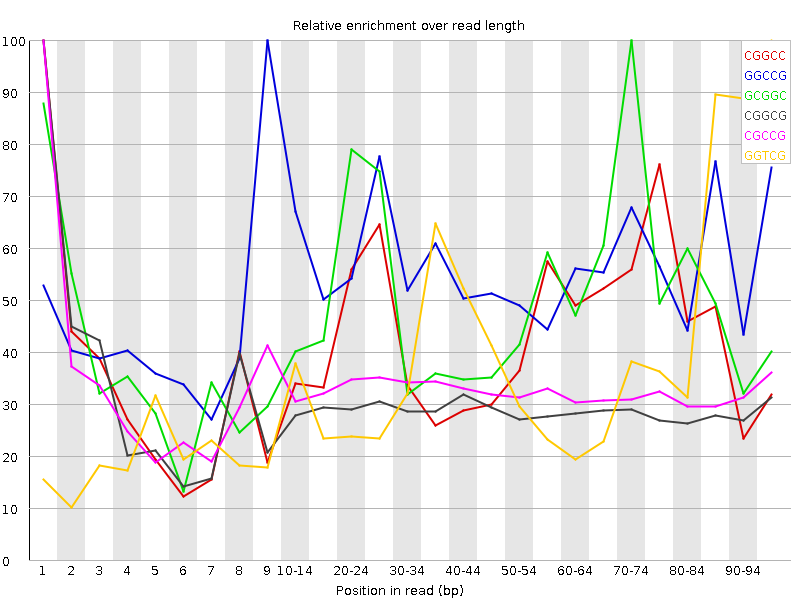
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1655070 | 3.5611677 | 8.273124 | 1 |
| GGCCG | 1594670 | 3.4547062 | 6.1987696 | 9 |
| GCGGC | 1496260 | 3.2415104 | 6.5052857 | 70-74 |
| CGGCG | 1216570 | 2.6355875 | 8.981237 | 1 |
| CGCCG | 1203230 | 2.5889564 | 7.918634 | 1 |
| GGTCG | 1865740 | 2.5715625 | 6.527176 | 95-97 |
| CACCG | 1883245 | 2.5614655 | 13.624136 | 1 |
| GCCGG | 1124535 | 2.436202 | 5.60637 | 1 |
| CTGGC | 1774850 | 2.4296484 | 10.66466 | 1 |
| CCGGC | 1129085 | 2.4294207 | 8.540261 | 1 |
| CCTCG | 1782570 | 2.4236174 | 6.6448636 | 1 |
| CGATC | 2779305 | 2.4050472 | 12.459572 | 4 |
| TGGCC | 1622125 | 2.2205784 | 5.198074 | 8 |
| GACCT | 2402935 | 2.0793586 | 8.328574 | 95-97 |
| CGACG | 1463350 | 2.0039835 | 5.8259873 | 1 |
| CCGCG | 927300 | 1.9952455 | 5.845528 | 1 |
| CGCGG | 916220 | 1.9849068 | 5.9315844 | 1 |
| CCGAT | 2281430 | 1.9742151 | 10.89616 | 3 |
| AGGTC | 2201115 | 1.9177604 | 5.5561857 | 95-97 |
| GGGGC | 866810 | 1.8907256 | 9.716423 | 95-97 |
| ACCGA | 2102040 | 1.8196696 | 8.938548 | 2 |
| CAGCG | 1315680 | 1.801757 | 7.0937147 | 1 |
| GGCTT | 2045240 | 1.781278 | 7.131742 | 3 |
| GCCAG | 1273265 | 1.7436718 | 5.2486897 | 1 |
| GTCGC | 1206190 | 1.6511917 | 7.3987045 | 95-97 |
| TGGCT | 1817670 | 1.5830786 | 5.753543 | 45-49 |
| CTTTC | 2796620 | 1.5286148 | 8.701222 | 1 |
| CGAAC | 1758935 | 1.5226543 | 5.5970683 | 90-94 |
| GATCT | 2755635 | 1.5171018 | 7.807658 | 5 |
| TCCTT | 2722630 | 1.4881723 | 7.0857954 | 2 |
| CCTTT | 2566105 | 1.4026167 | 7.0827045 | 3 |
| AGACC | 1532470 | 1.3266107 | 6.4173894 | 95-97 |
| CAGAC | 1483300 | 1.2840458 | 5.861175 | 95-97 |
| CAAAT | 3596490 | 1.2521077 | 5.216538 | 9 |
| GCGTA | 1415365 | 1.2331618 | 5.840808 | 1 |
| CTCGG | 895215 | 1.2254882 | 5.129092 | 1 |
| ACACC | 1424650 | 1.2248855 | 5.8058643 | 8 |
| CTAAA | 3336550 | 1.1616102 | 5.151899 | 8 |
| CGTAC | 1321880 | 1.1438773 | 10.660875 | 5 |
| TTTCG | 2064045 | 1.1359208 | 7.32338 | 2 |
| TTCGT | 2045460 | 1.1256927 | 7.33505 | 6 |
| CTCCC | 803945 | 1.0856245 | 5.070389 | 1 |
| CCGGG | 488155 | 1.0575432 | 5.3077455 | 1 |
| CTCAT | 1907930 | 1.0432566 | 5.1498647 | 1 |
| GTCCT | 1080890 | 0.93498504 | 9.908045 | 1 |
| TCGTA | 1653850 | 0.9105193 | 7.145583 | 7 |
| GTACA | 1594305 | 0.87806886 | 7.323728 | 6 |
| TACAA | 2192935 | 0.763464 | 5.2508254 | 7 |