![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-24-R2_val_2.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 17868629 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 38 |
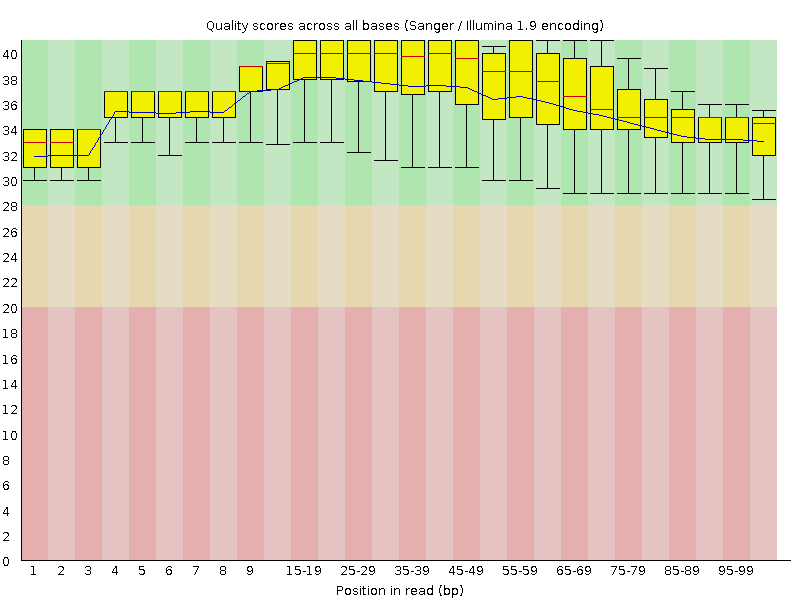
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
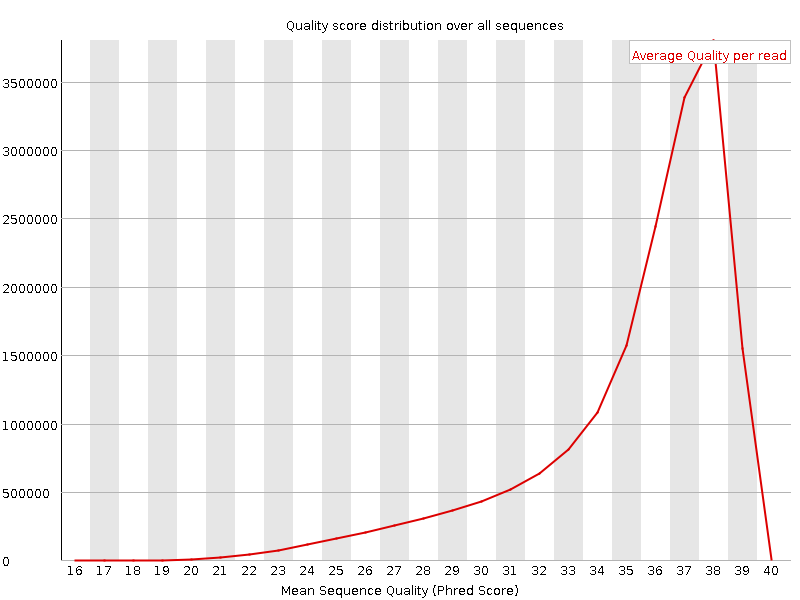
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
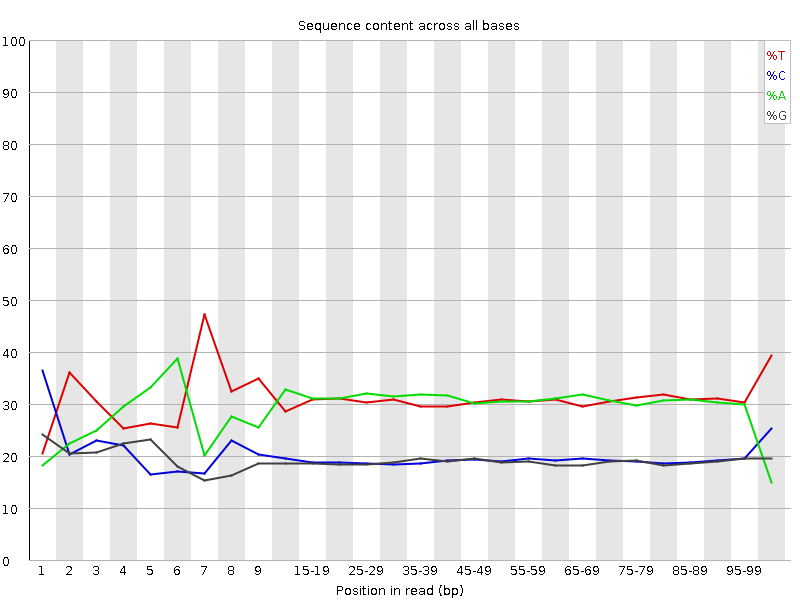
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
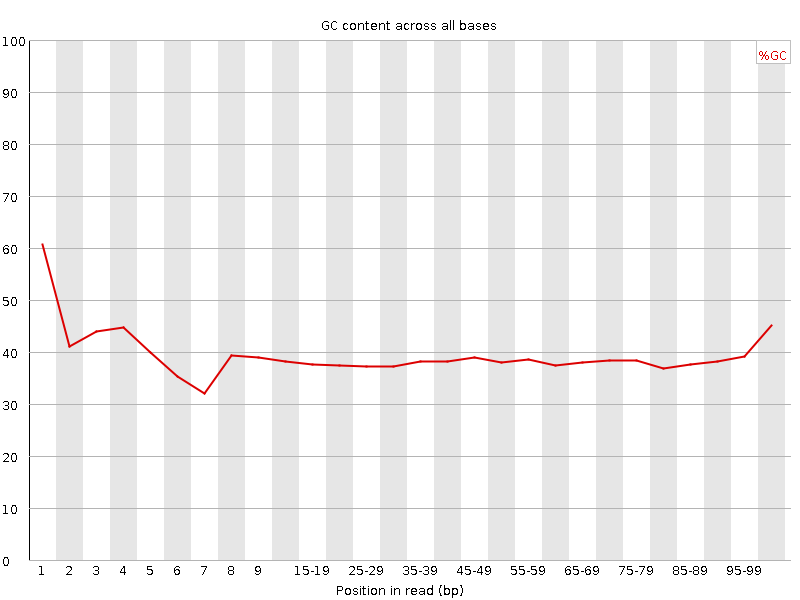
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
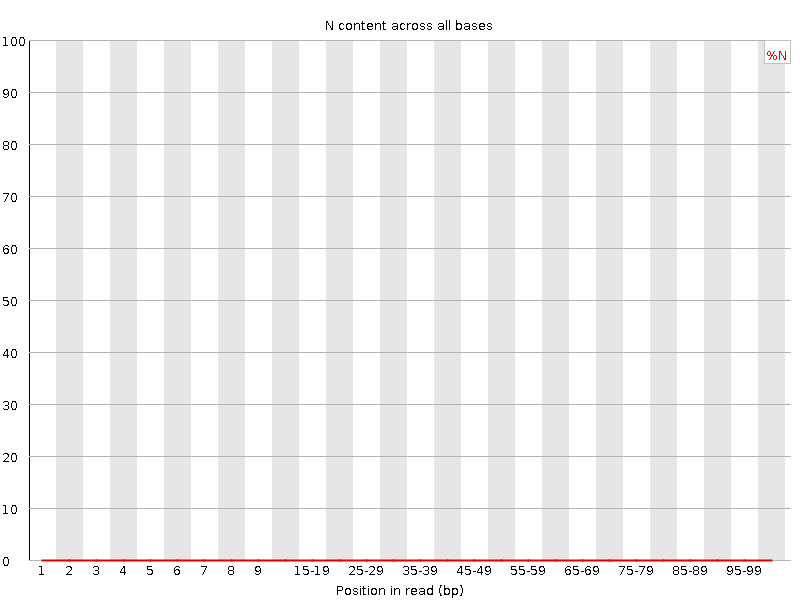
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
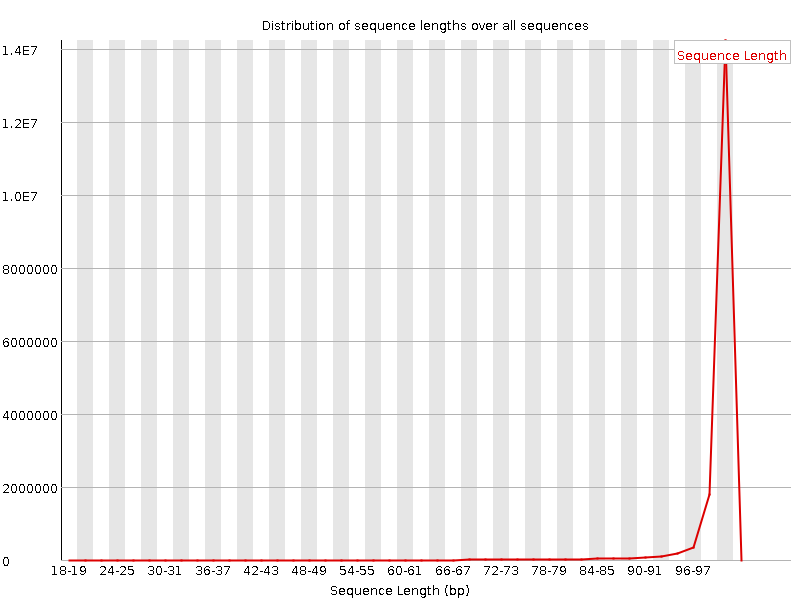
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 123698 | 0.6922635194899396 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 118437 | 0.6628208577166161 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 88014 | 0.492561572575042 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 70542 | 0.3947812672141774 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 54286 | 0.3038061845707357 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 47558 | 0.2661536036144687 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 38528 | 0.21561810925729108 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 34196 | 0.19137450332647232 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 30689 | 0.17174792761101035 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 30179 | 0.16889376347788068 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 27918 | 0.15624030248767268 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 27207 | 0.1522612619020743 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 26211 | 0.14668724724207996 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 25867 | 0.14476208555228273 | No Hit |
| TTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCTT | 25736 | 0.14402895711808666 | No Hit |
| CCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTAAA | 24648 | 0.1379400736340768 | No Hit |
| CCTCGATGTTGAATTAAAATAAAAATTAGATGTAGATGTCTAATATTTAG | 22805 | 0.12762590795298284 | No Hit |
| TTATAATTAAGAAAGAATTAATTACCTTAGGGATAACAGCGTAATATTTT | 22772 | 0.12744122674436859 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 22069 | 0.12350695736085852 | No Hit |
| CAAGTTTTTAATTAAAAAACAATTGATTATGCTACCTTTGCACAGTCATT | 21293 | 0.11916415075829265 | No Hit |
| GTCTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATT | 20808 | 0.11644989663168898 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 20308 | 0.11365169650116973 | No Hit |
| AATTAATTACCTTAGGGATAACAGCGTAATATTTTTAGAAAGATCATATT | 19306 | 0.10804410343960916 | No Hit |
| GTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATG | 19143 | 0.10713189019705988 | No Hit |
| CCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGA | 19135 | 0.10708711899497159 | No Hit |
| GCTACCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGG | 18910 | 0.10582792893623792 | No Hit |
| GGCAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATA | 18843 | 0.10545297011874832 | No Hit |
| CTCCCAACTAAATATAATTCATATTATTAATGATACAAAAATTTTTAATA | 18039 | 0.10095346430887338 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content

| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1631275 | 3.5797682 | 8.503526 | 1 |
| GGCCG | 1554965 | 3.4918883 | 5.7822504 | 9 |
| GCGGC | 1480295 | 3.3242066 | 6.853418 | 70-74 |
| GGTCG | 1940085 | 2.7599225 | 7.4956183 | 95-97 |
| CACCG | 1929980 | 2.6425314 | 15.074572 | 1 |
| CGGCG | 1172955 | 2.6340322 | 9.788511 | 1 |
| CGCCG | 1180820 | 2.5912623 | 8.750408 | 1 |
| CTGGC | 1811495 | 2.5182638 | 11.143438 | 1 |
| CGATC | 2890945 | 2.5075154 | 13.642109 | 4 |
| GCCGG | 1088315 | 2.4439611 | 5.804396 | 1 |
| CCGGC | 1095035 | 2.4030106 | 8.31139 | 1 |
| CGCGC | 1071155 | 2.350607 | 5.1101723 | 1 |
| CCTCG | 1690480 | 2.2964773 | 6.940134 | 1 |
| CCGAT | 2406640 | 2.0874445 | 11.954251 | 3 |
| GACCT | 2350985 | 2.039171 | 8.912602 | 95-97 |
| CGCGG | 889625 | 1.9977754 | 6.3329334 | 1 |
| GGGGC | 868245 | 1.9952348 | 10.81567 | 95-97 |
| CGACG | 1418030 | 1.9868486 | 5.787519 | 1 |
| CCGCG | 903235 | 1.9821131 | 6.1627827 | 1 |
| AGGTC | 2231000 | 1.9802289 | 5.9749284 | 95-97 |
| ACCGA | 2226040 | 1.946041 | 9.89032 | 2 |
| CGAGG | 1321350 | 1.8945639 | 5.2154922 | 90-94 |
| GGCTT | 2092195 | 1.8424797 | 7.191181 | 3 |
| CCGCC | 822950 | 1.7647741 | 5.03409 | 1 |
| CAGCG | 1253670 | 1.7565585 | 6.8408155 | 1 |
| GCCAG | 1235490 | 1.7310858 | 5.323858 | 1 |
| GTCGC | 1231130 | 1.7114649 | 8.584325 | 95-97 |
| TGGCT | 1857290 | 1.6356119 | 6.650035 | 45-49 |
| GATCT | 2895325 | 1.5908805 | 8.45814 | 5 |
| CGAAC | 1815715 | 1.5873281 | 6.5395036 | 90-94 |
| CTTTC | 2939420 | 1.5659384 | 9.471406 | 1 |
| TCCTT | 2849520 | 1.5180451 | 8.314745 | 2 |
| CCTTT | 2652960 | 1.4133302 | 8.320911 | 3 |
| AGACC | 1567660 | 1.3704745 | 7.421843 | 95-97 |
| CAGAC | 1526395 | 1.3343998 | 6.764872 | 95-97 |
| CAAAT | 3739640 | 1.2921858 | 5.742074 | 9 |
| ACACC | 1486535 | 1.269937 | 5.9628716 | 8 |
| ATCTA | 3660365 | 1.254886 | 5.02016 | 6 |
| CTCGG | 888560 | 1.2352387 | 5.58448 | 1 |
| CTCGT | 1431950 | 1.2322999 | 5.093637 | 1 |
| GCGTA | 1355145 | 1.2028226 | 5.4140186 | 1 |
| CGTAC | 1378210 | 1.1954163 | 12.3350115 | 8 |
| CTAAA | 3444400 | 1.1901692 | 5.6123185 | 8 |
| TTTCG | 2145905 | 1.1698642 | 8.682289 | 5 |
| TTCGT | 2134530 | 1.163663 | 8.671333 | 6 |
| CTCCC | 814235 | 1.0809109 | 5.7246504 | 1 |
| CCGGG | 473690 | 1.0637361 | 5.4249067 | 1 |
| CTCAT | 1967485 | 1.0564275 | 5.2109833 | 1 |
| GTCCT | 1122295 | 0.96581864 | 12.048676 | 1 |
| TCGTA | 1715170 | 0.9424263 | 8.538967 | 7 |
| GGGTC | 643135 | 0.91490984 | 5.2997355 | 2 |
| GTACA | 1639875 | 0.908168 | 8.249553 | 6 |
| ACAAA | 2415435 | 0.84121263 | 5.323141 | 8 |
| TACAA | 2275820 | 0.78638107 | 5.757001 | 7 |