![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-28-R1_val_1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 13905764 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 39 |
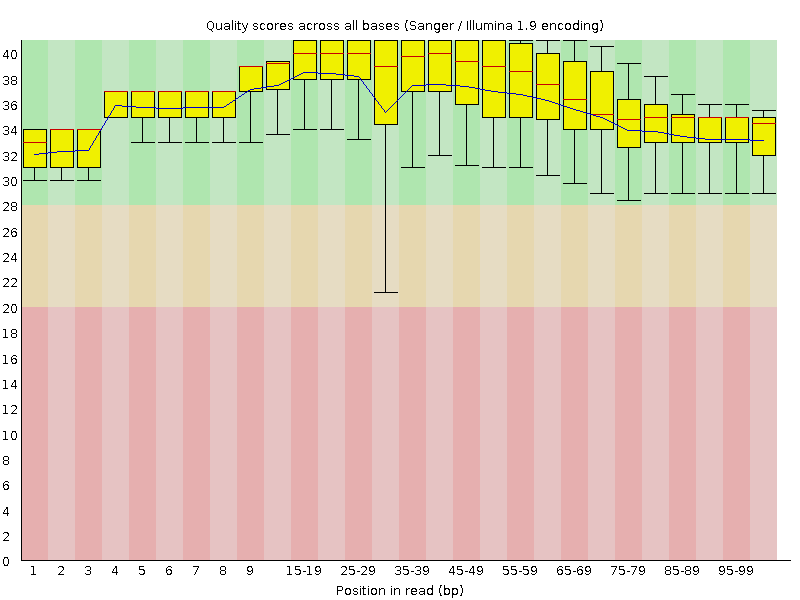
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
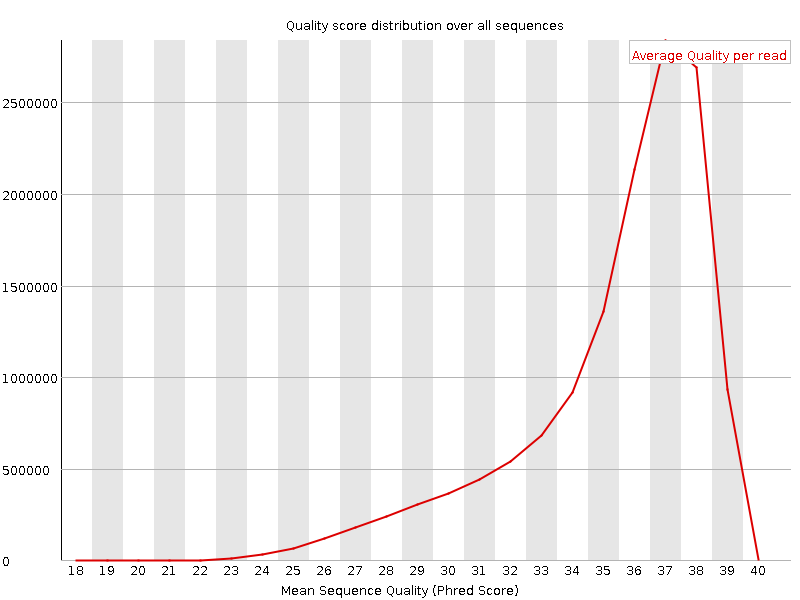
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
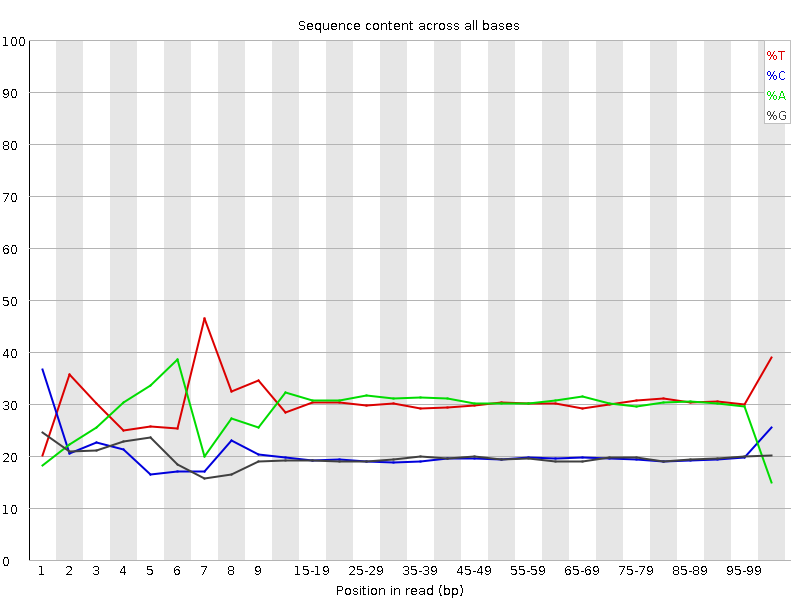
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
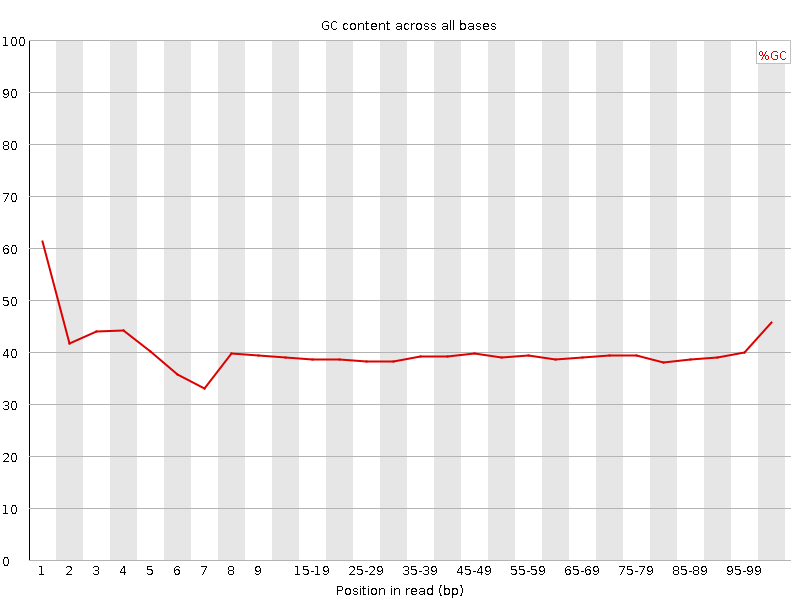
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution

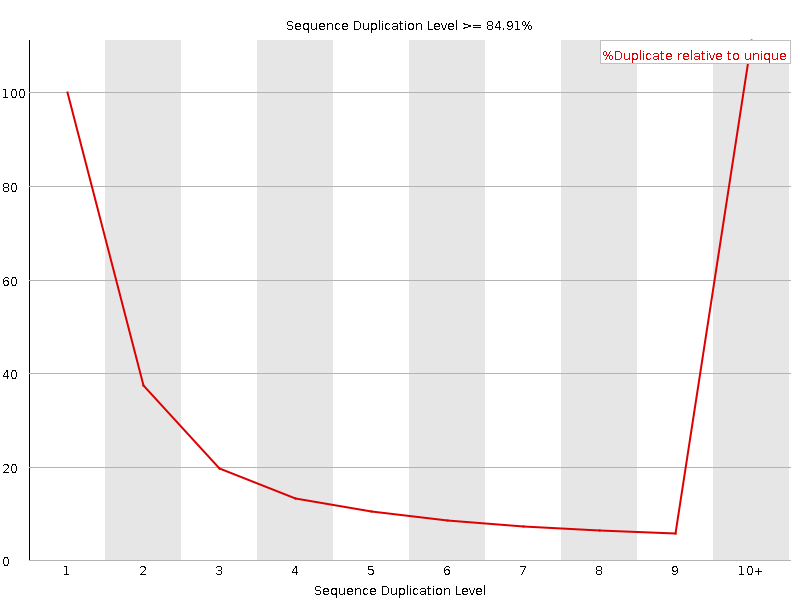
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 66821 | 0.4805273554189472 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 55878 | 0.4018333692417044 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 46705 | 0.33586791779293823 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 42451 | 0.3052762868692436 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 29824 | 0.21447221454355186 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 28340 | 0.2038003809067952 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 28337 | 0.2037788071191198 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 22142 | 0.15922893556945164 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 20276 | 0.1458100396353627 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 20044 | 0.14414166672179968 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 18153 | 0.13054298922374924 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 16987 | 0.12215797708058328 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 14986 | 0.10776826070110208 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 14895 | 0.1071138558082821 | No Hit |
| CAAGTTTTTAATTAAAAAACAATTGATTATGCTACCTTTGCACAGTCATT | 14464 | 0.10401442164558523 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 14179 | 0.10196491181642375 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 14047 | 0.10101566515870684 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
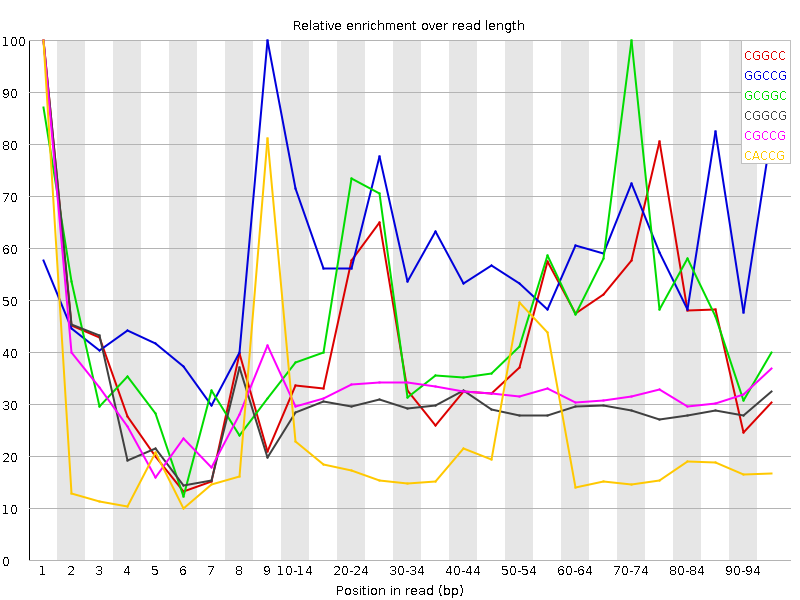
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1279400 | 3.2466478 | 7.4154534 | 1 |
| GGCCG | 1232345 | 3.1582327 | 5.313756 | 9 |
| GCGGC | 1195075 | 3.062718 | 6.314579 | 70-74 |
| CGGCG | 977490 | 2.5050948 | 8.369431 | 1 |
| CGCCG | 970955 | 2.4639275 | 7.569942 | 1 |
| CACCG | 1426955 | 2.3530715 | 10.821159 | 1 |
| GGTCG | 1366115 | 2.2904727 | 5.3213024 | 95-97 |
| CGATC | 2105945 | 2.2719455 | 10.790951 | 4 |
| GCCGG | 863530 | 2.2130399 | 5.3336973 | 1 |
| CCTCG | 1343355 | 2.2083228 | 6.245189 | 1 |
| CCGGC | 865180 | 2.1955094 | 7.462159 | 1 |
| CTGGC | 1318375 | 2.1887379 | 8.89103 | 1 |
| CGACG | 1186020 | 1.9751492 | 5.5604806 | 1 |
| GGCTT | 1791705 | 1.9460198 | 6.0130386 | 3 |
| CCGAT | 1768560 | 1.9079661 | 9.265651 | 3 |
| CGCGG | 725480 | 1.8592478 | 5.576798 | 1 |
| CCGCG | 731175 | 1.8554538 | 5.4921303 | 1 |
| GACCT | 1711415 | 1.8463167 | 7.386073 | 95-97 |
| GCCGA | 1087785 | 1.8115528 | 5.432373 | 1 |
| TGGCT | 1637730 | 1.7787833 | 5.3412404 | 45-49 |
| GCAAA | 2468030 | 1.7528046 | 5.059435 | 2 |
| ACCGA | 1580845 | 1.7107763 | 7.2679105 | 2 |
| GGGGC | 659040 | 1.7057158 | 9.278116 | 95-97 |
| CAGCG | 1010535 | 1.6829036 | 6.2442427 | 1 |
| ATATT | 5432830 | 1.6301405 | 5.089228 | 4 |
| GTCGC | 928635 | 1.5417 | 5.971051 | 95-97 |
| CTTTC | 2099180 | 1.4624816 | 8.329896 | 1 |
| GATCT | 2064125 | 1.4568433 | 6.870156 | 5 |
| TCCTT | 2069910 | 1.4420894 | 6.5048747 | 2 |
| CGAAC | 1294985 | 1.4014212 | 5.0885544 | 90-94 |
| CCTTT | 1960200 | 1.3656553 | 6.5071726 | 3 |
| CTAAA | 2947320 | 1.35598 | 5.6979938 | 8 |
| CAAAT | 2711185 | 1.2473408 | 5.003518 | 9 |
| GCGTA | 1128885 | 1.2299391 | 5.084173 | 1 |
| AGACC | 1126770 | 1.2193803 | 5.813542 | 95-97 |
| CTCGG | 725710 | 1.2048082 | 5.3034353 | 1 |
| CAGAC | 1069755 | 1.1576793 | 5.376628 | 95-97 |
| TTTCG | 1585175 | 1.1153238 | 6.9346385 | 2 |
| TTCGT | 1563890 | 1.1003478 | 6.8202767 | 3 |
| CGTAC | 1015375 | 1.0954115 | 9.783063 | 5 |
| CTCAT | 1561155 | 1.091038 | 5.556082 | 1 |
| CTCCC | 621460 | 1.0115838 | 5.1760406 | 1 |
| GTCCT | 851225 | 0.9154659 | 8.751115 | 1 |
| TCGTA | 1275635 | 0.9003332 | 6.496738 | 7 |
| GTACA | 1210780 | 0.8572257 | 6.964643 | 6 |
| TACAA | 1691835 | 0.77836627 | 5.113195 | 7 |