![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-31-R1_val_1.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 14239058 |
| Filtered Sequences | 0 |
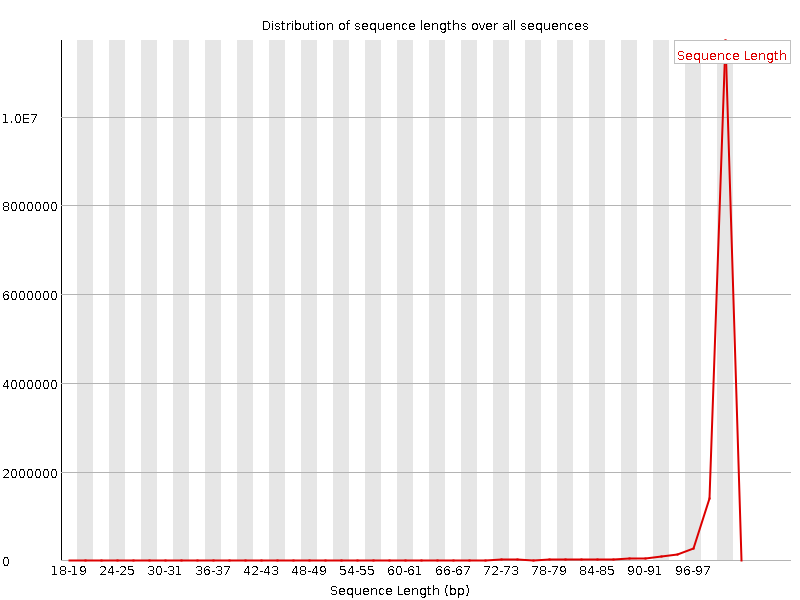
| Sequence length | 20-101 |
| %GC | 39 |
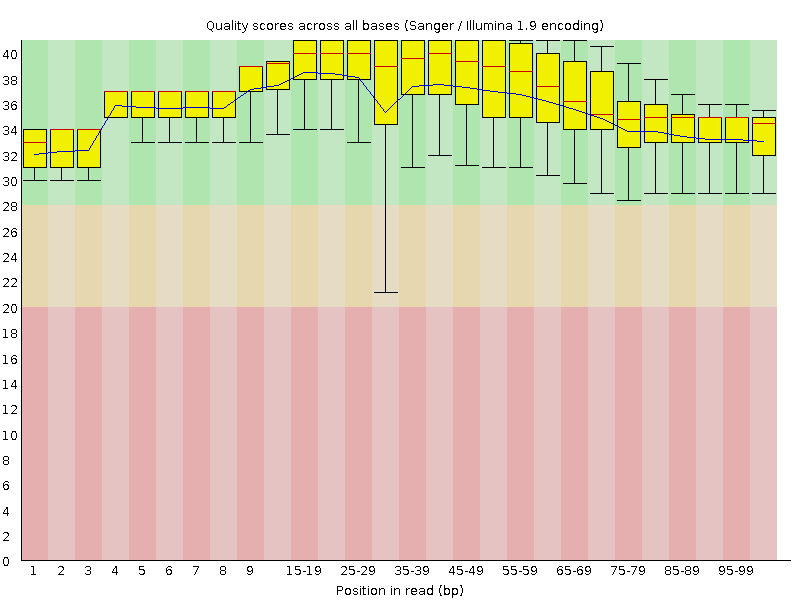
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
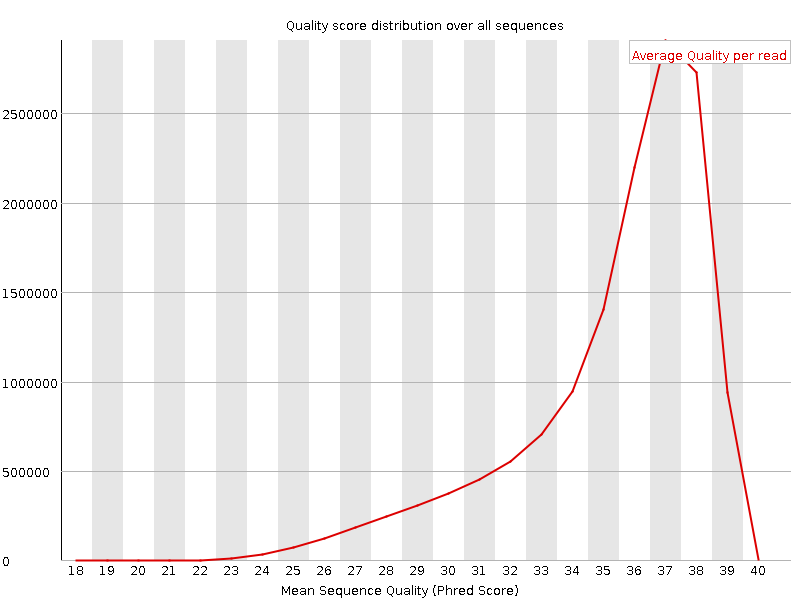
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content

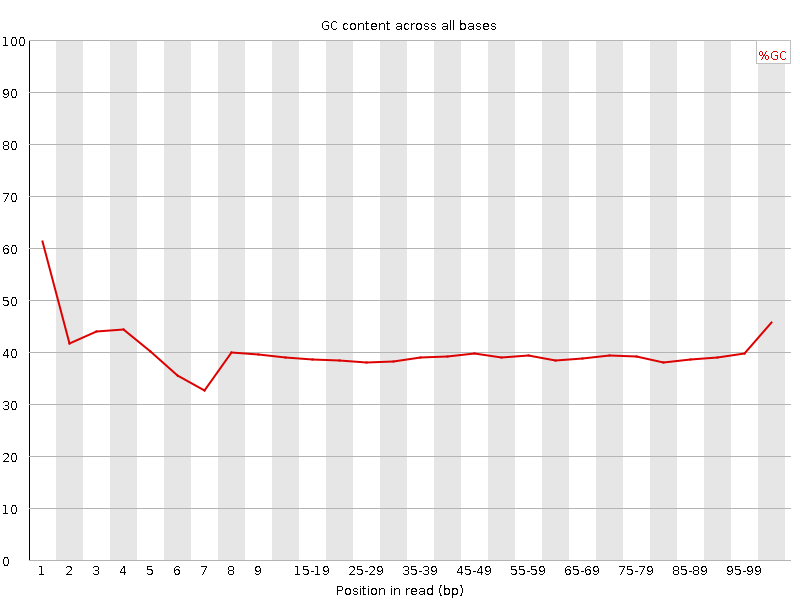
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
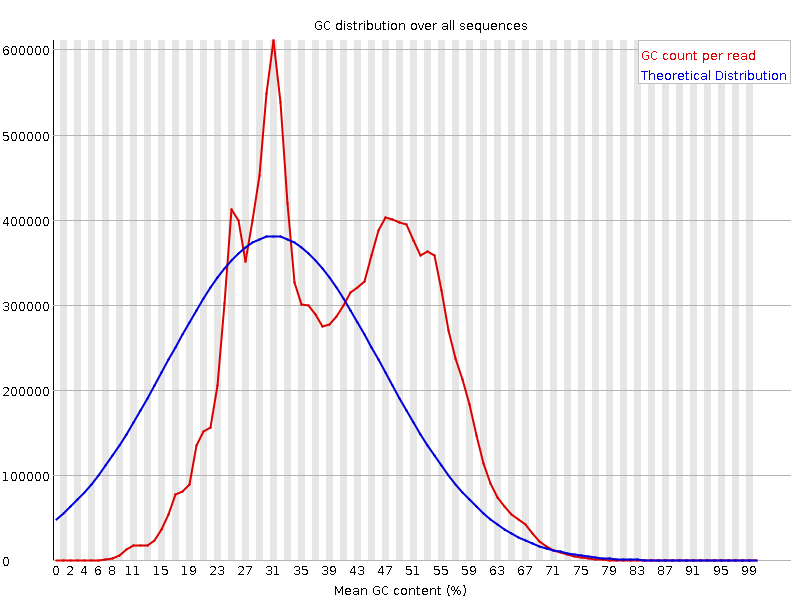
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content

![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
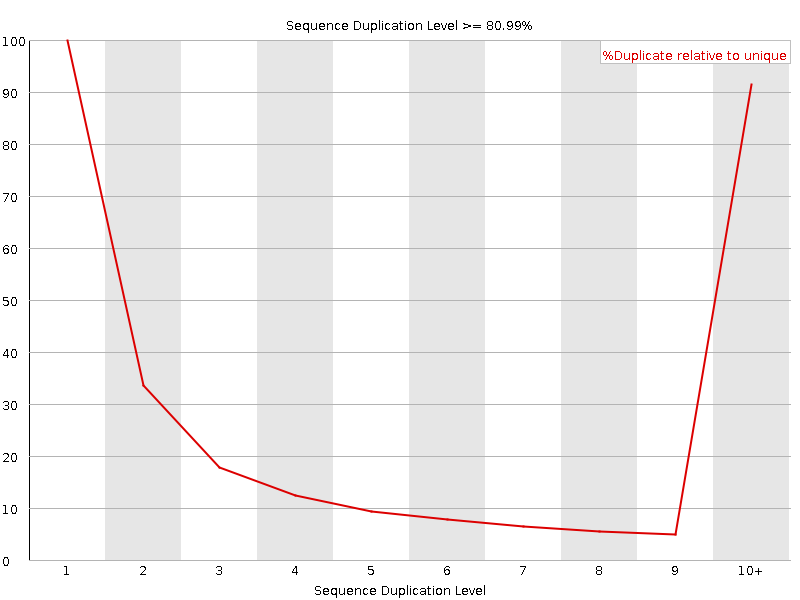
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 65669 | 0.4611892163091126 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 56929 | 0.3998087513935262 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 47221 | 0.33163008395639654 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 42654 | 0.29955633301023143 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 29689 | 0.20850396142778546 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 29516 | 0.2072889934151543 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 28932 | 0.20318759850546292 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 21248 | 0.1492233545224691 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 20300 | 0.14256561073070986 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 19545 | 0.13726329368136572 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 19294 | 0.13550053662257713 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 18050 | 0.12676400363001541 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 17698 | 0.12429192998581788 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 16023 | 0.11252851136641202 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 15843 | 0.11126438279835647 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 15460 | 0.10857459812299382 | No Hit |
| CAAGTTTTTAATTAAAAAACAATTGATTATGCTACCTTTGCACAGTCATT | 14982 | 0.1052176344811574 | No Hit |
| GCTACCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGG | 14486 | 0.10173425798251541 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
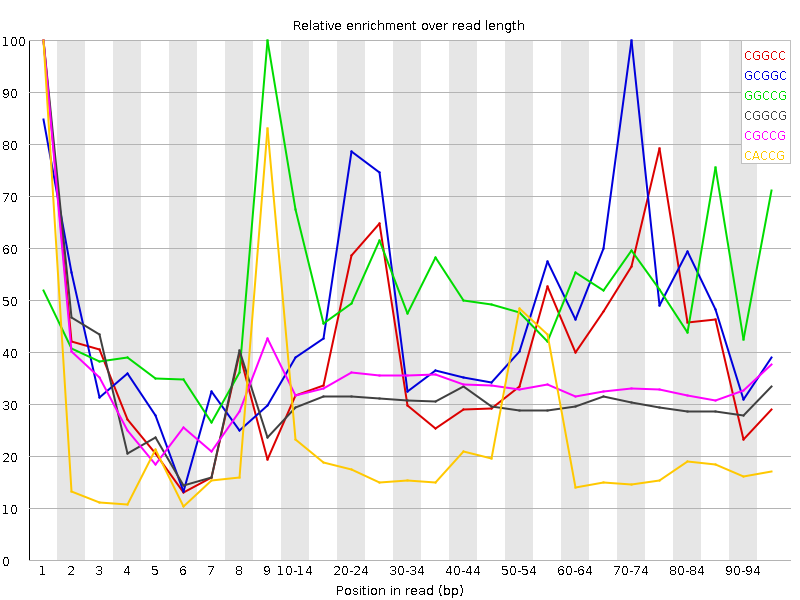
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1273245 | 3.180394 | 7.6219873 | 1 |
| GCGGC | 1242870 | 3.128366 | 6.3483005 | 70-74 |
| GGCCG | 1182000 | 2.9751532 | 5.653399 | 9 |
| CGGCG | 1019600 | 2.5663843 | 8.30305 | 1 |
| CGCCG | 999185 | 2.495829 | 7.355542 | 1 |
| CACCG | 1447190 | 2.3460312 | 10.820562 | 1 |
| CGATC | 2150610 | 2.273427 | 10.732451 | 4 |
| GGTCG | 1383915 | 2.271498 | 5.2027445 | 95-97 |
| CCTCG | 1403780 | 2.269113 | 6.1683335 | 1 |
| CTGGC | 1304285 | 2.124479 | 9.065635 | 1 |
| CGACG | 1270285 | 2.0750682 | 5.787998 | 1 |
| CCGGC | 819605 | 2.0472624 | 6.9228625 | 1 |
| GCCGG | 810275 | 2.0395029 | 5.0775843 | 1 |
| CGCGG | 761890 | 1.9177153 | 5.7939587 | 1 |
| CCGCG | 759105 | 1.8961416 | 5.7049956 | 1 |
| CCGAT | 1746285 | 1.8460116 | 8.965292 | 3 |
| GACCT | 1692220 | 1.7888594 | 7.0961924 | 95-97 |
| ACCGA | 1633105 | 1.731349 | 7.151468 | 2 |
| CAGCG | 1047785 | 1.7116041 | 6.193551 | 1 |
| GGGGC | 665430 | 1.6877851 | 8.966013 | 95-97 |
| GGCTT | 1527010 | 1.6219332 | 5.787745 | 3 |
| GTCGC | 987240 | 1.6080617 | 5.9346614 | 95-97 |
| TGGCT | 1428810 | 1.5176287 | 5.0428543 | 45-49 |
| CTTTC | 2223240 | 1.5165007 | 8.144665 | 1 |
| GATCT | 2122690 | 1.4632447 | 6.9576573 | 5 |
| TCCTT | 2082130 | 1.4202477 | 6.3827543 | 2 |
| CCTTT | 2020695 | 1.3783422 | 6.496526 | 3 |
| CTCGG | 788165 | 1.2837993 | 5.4538345 | 1 |
| ACACC | 1136265 | 1.1954385 | 5.0710263 | 8 |
| CAGAC | 1115315 | 1.1824099 | 5.2526455 | 95-97 |
| AGACC | 1110745 | 1.1775651 | 5.543226 | 95-97 |
| TTTCG | 1649155 | 1.1335502 | 6.6471434 | 2 |
| CGTAC | 1048135 | 1.1079919 | 9.441948 | 5 |
| TTCGT | 1608515 | 1.1056162 | 6.567475 | 6 |
| CTCAT | 1606730 | 1.0991333 | 5.4446893 | 1 |
| GTCCT | 912245 | 0.96156716 | 8.739699 | 1 |
| CCGGG | 377165 | 0.9493431 | 5.006303 | 1 |
| GTACA | 1323515 | 0.9149776 | 6.8031416 | 6 |
| TCGTA | 1316460 | 0.90748215 | 6.4331217 | 7 |
| TACAA | 1810760 | 0.8124223 | 5.057189 | 7 |