![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-31-R2_val_2.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 14239058 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 39 |
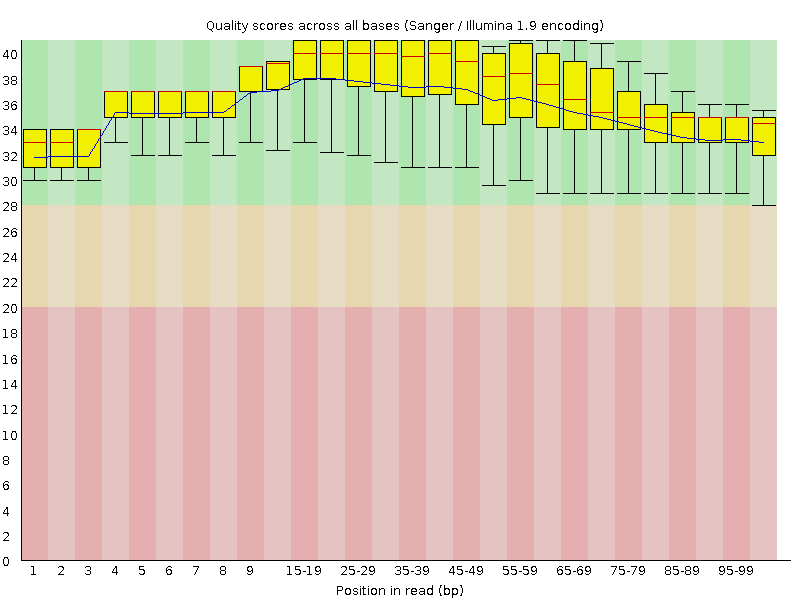
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
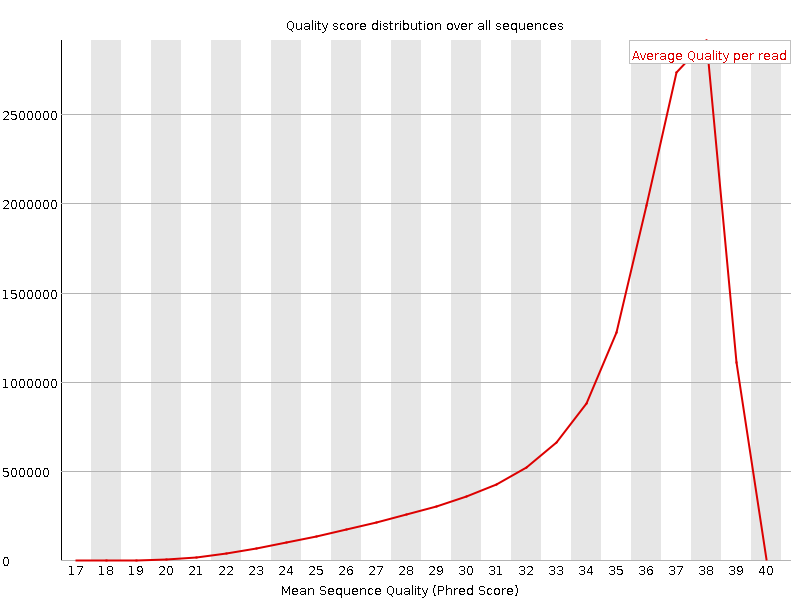
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
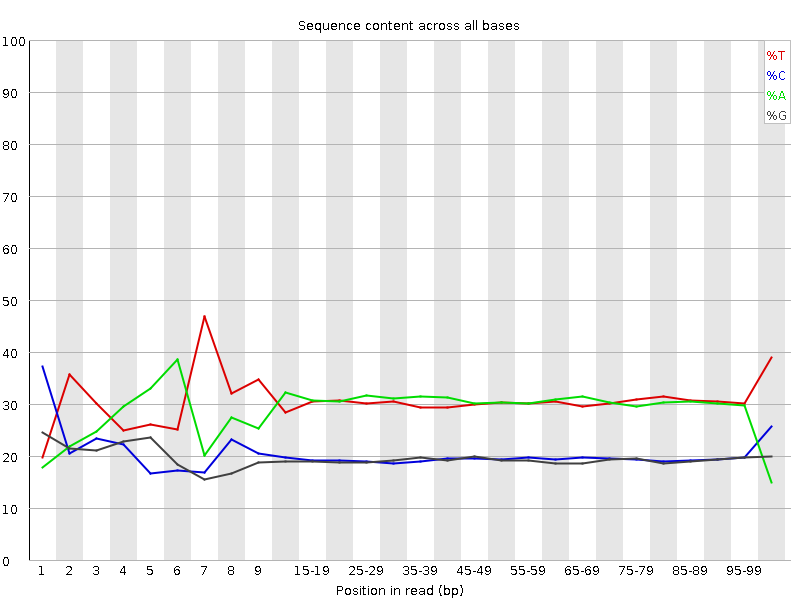
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
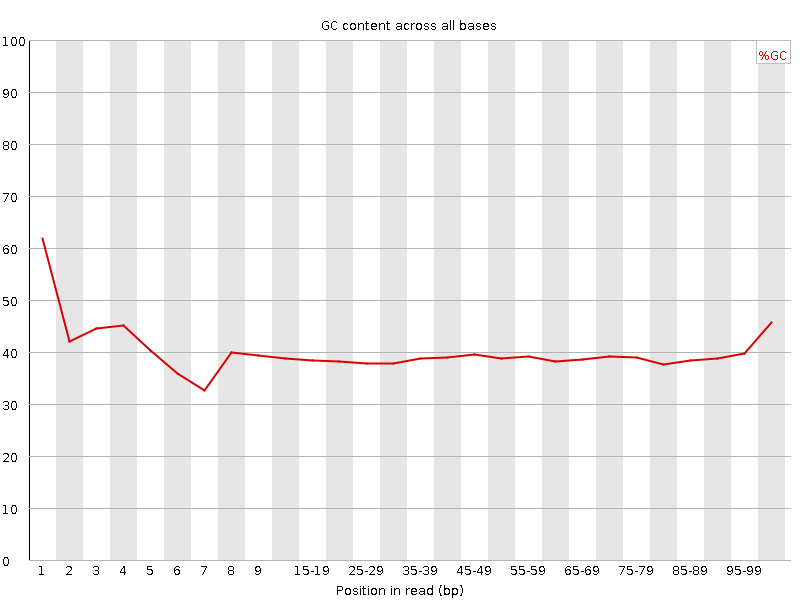
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content

![[OK]](Icons/tick.png) Per base N content
Per base N content
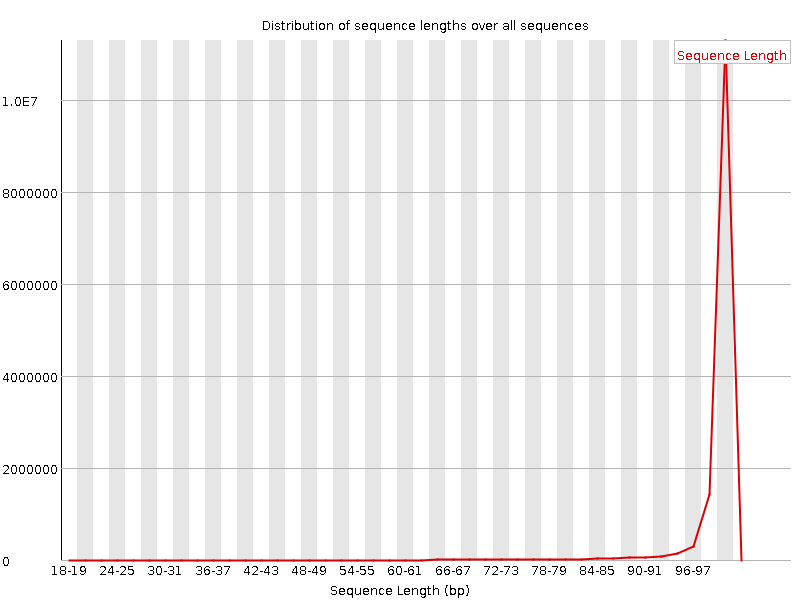
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
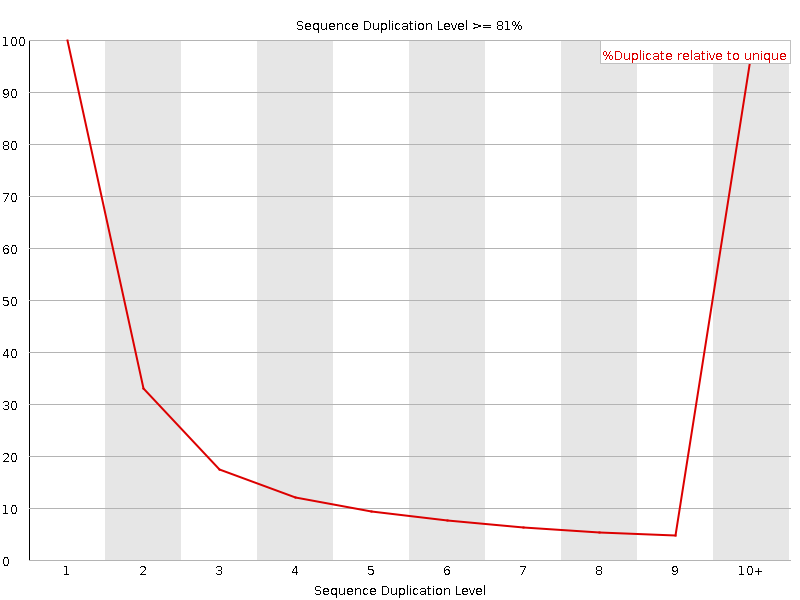
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 89175 | 0.6262703614241898 | No Hit |
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 83609 | 0.5871806969253163 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 59390 | 0.4170921980934413 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 58016 | 0.4074426833572839 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 35901 | 0.25213044289868053 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 31566 | 0.2216860132180092 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 30583 | 0.21478246664912806 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 22830 | 0.1603336400483796 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 22812 | 0.16020722719157404 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 22405 | 0.15734889204047064 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 22326 | 0.15679408005782405 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 22288 | 0.15652720847123452 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 20773 | 0.14588745969010028 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 19444 | 0.13655397709595676 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 19439 | 0.13651886241351077 | No Hit |
| CAAGTTTTTAATTAAAAAACAATTGATTATGCTACCTTTGCACAGTCATT | 17371 | 0.12199542975385028 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 17345 | 0.12181283340513116 | No Hit |
| GTCTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATT | 17005 | 0.11942503499880398 | No Hit |
| GCTACCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGG | 16871 | 0.11848396150925151 | No Hit |
| CCTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGA | 16695 | 0.11724792468715277 | No Hit |
| TTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCTT | 16554 | 0.1162576906421759 | No Hit |
| CCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTAAA | 15954 | 0.11204392874865739 | No Hit |
| CCTCGATGTTGAATTAAAATAAAAATTAGATGTAGATGTCTAATATTTAG | 15625 | 0.10973338264371141 | No Hit |
| GGCAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATA | 14945 | 0.10495778583105707 | No Hit |
| AAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTG | 14718 | 0.10336357924800924 | No Hit |
| CTTAAACCTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTG | 14673 | 0.10304754710599535 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
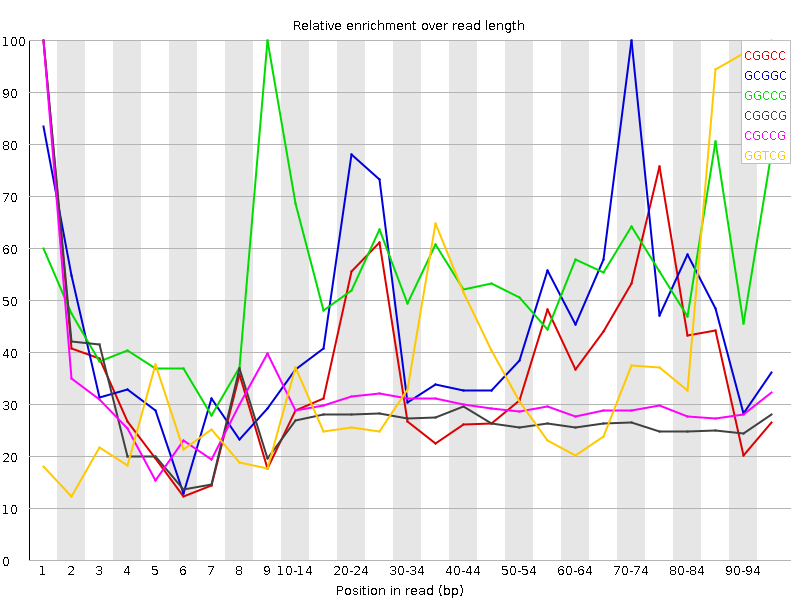
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1266735 | 3.2221618 | 8.272162 | 1 |
| GCGGC | 1230110 | 3.1938174 | 6.668001 | 70-74 |
| GGCCG | 1152910 | 2.9933774 | 5.384699 | 9 |
| CGGCG | 983755 | 2.5541894 | 9.29935 | 1 |
| CGCCG | 987830 | 2.512718 | 8.331965 | 1 |
| GGTCG | 1443700 | 2.4356718 | 6.0317326 | 95-97 |
| CACCG | 1486150 | 2.4258323 | 12.378875 | 1 |
| CGATC | 2230585 | 2.3658798 | 11.848117 | 4 |
| CTGGC | 1330720 | 2.1994998 | 9.283802 | 1 |
| CCTCG | 1331725 | 2.156489 | 6.5422845 | 1 |
| GCCGG | 792155 | 2.056725 | 5.10811 | 1 |
| CGACG | 1233735 | 2.0555336 | 5.874226 | 1 |
| CCGGC | 792920 | 2.0169303 | 7.1107574 | 1 |
| CCGAT | 1837235 | 1.9486713 | 10.201099 | 3 |
| CGCGG | 735525 | 1.9096929 | 6.0274234 | 1 |
| CCGCG | 741345 | 1.8857403 | 5.9469614 | 1 |
| ACCGA | 1717505 | 1.8362736 | 8.097897 | 2 |
| GGGGC | 670755 | 1.7776022 | 10.024398 | 95-97 |
| GACCT | 1645505 | 1.7453119 | 7.5986733 | 95-97 |
| GGCTT | 1564960 | 1.6808007 | 5.9408436 | 3 |
| GTCGC | 1009150 | 1.667988 | 7.106158 | 95-97 |
| CAGCG | 997560 | 1.6620411 | 5.7018695 | 1 |
| TGGCT | 1455655 | 1.5634046 | 5.837719 | 45-49 |
| CTTTC | 2316875 | 1.5519657 | 8.842729 | 1 |
| GATCT | 2218980 | 1.5293361 | 7.563816 | 5 |
| CGAAC | 1381930 | 1.4774928 | 5.8045173 | 90-94 |
| TCCTT | 2147185 | 1.4382983 | 7.5288863 | 2 |
| CCTTT | 2071825 | 1.3878182 | 7.5661097 | 3 |
| CTCGG | 781350 | 1.2914656 | 5.849301 | 1 |
| CAAAT | 2878140 | 1.2831093 | 5.399428 | 9 |
| CTCGT | 1207010 | 1.2700454 | 5.118979 | 1 |
| ACACC | 1181605 | 1.2376765 | 5.160538 | 8 |
| CAGAC | 1147335 | 1.2266752 | 6.065303 | 95-97 |
| AGACC | 1134080 | 1.2125036 | 6.3698764 | 95-97 |
| CTAAA | 2692480 | 1.2003399 | 5.348055 | 8 |
| TTTCG | 1694110 | 1.1583123 | 7.712798 | 5 |
| CGTAC | 1079765 | 1.1452576 | 10.742098 | 8 |
| TTCGT | 1666295 | 1.1392944 | 7.7454 | 6 |
| CTCAT | 1657735 | 1.119335 | 5.605511 | 1 |
| CGGGG | 392320 | 1.0397073 | 5.090556 | 1 |
| CTCCC | 628190 | 0.9965959 | 5.347226 | 1 |
| GTCCT | 937495 | 0.98645514 | 10.595188 | 1 |
| GTACA | 1351800 | 0.9391339 | 7.6506734 | 6 |
| TCGTA | 1354880 | 0.93379265 | 7.704659 | 7 |
| GGGTC | 526140 | 0.8876529 | 5.456369 | 2 |
| ACAAA | 1909745 | 0.8582081 | 5.1016498 | 8 |
| TACAA | 1864315 | 0.8311339 | 5.5842657 | 7 |