![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-35-R2_val_2.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 15554492 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 39 |
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality

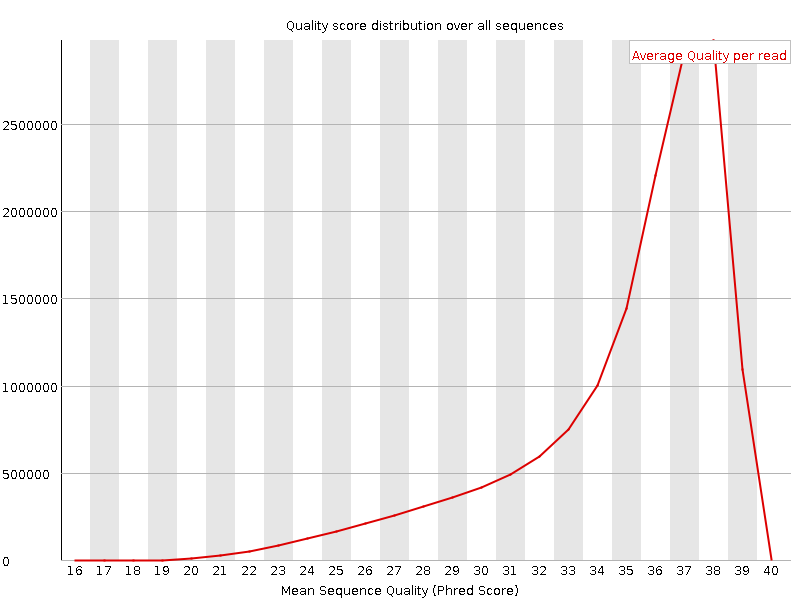
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
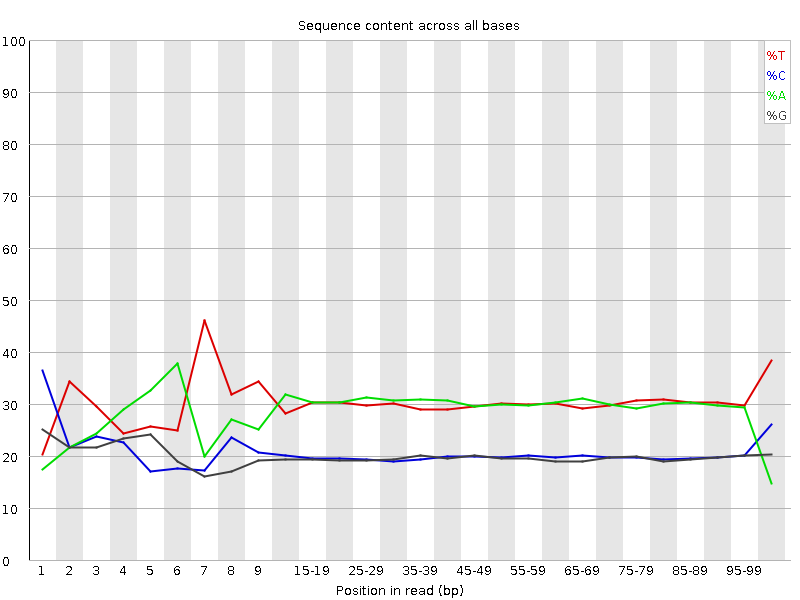
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
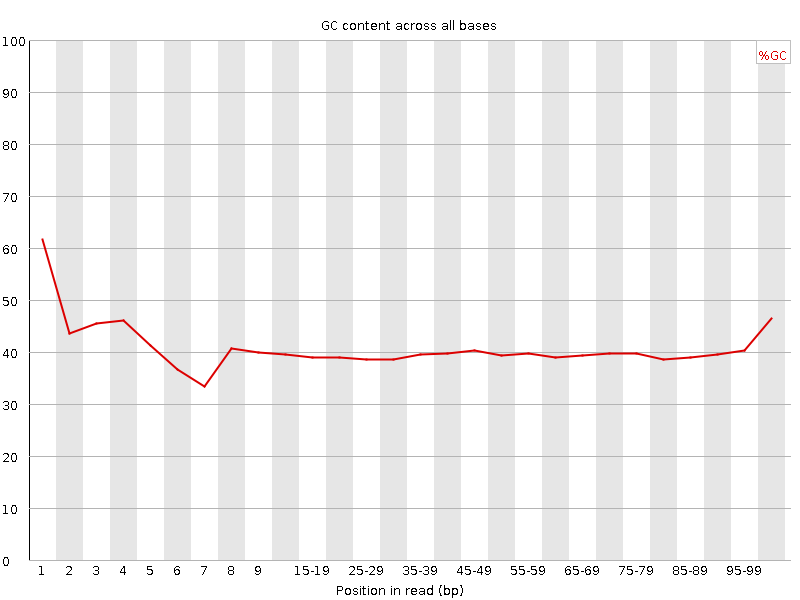
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
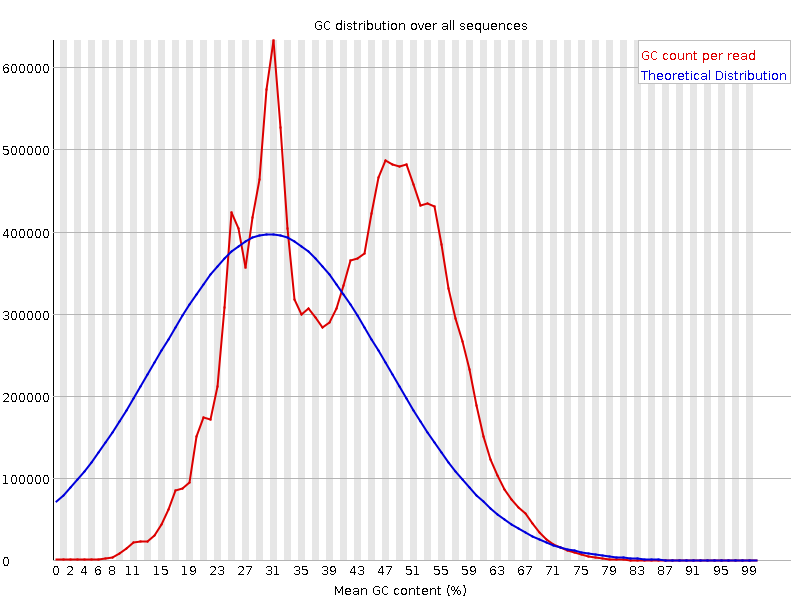
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
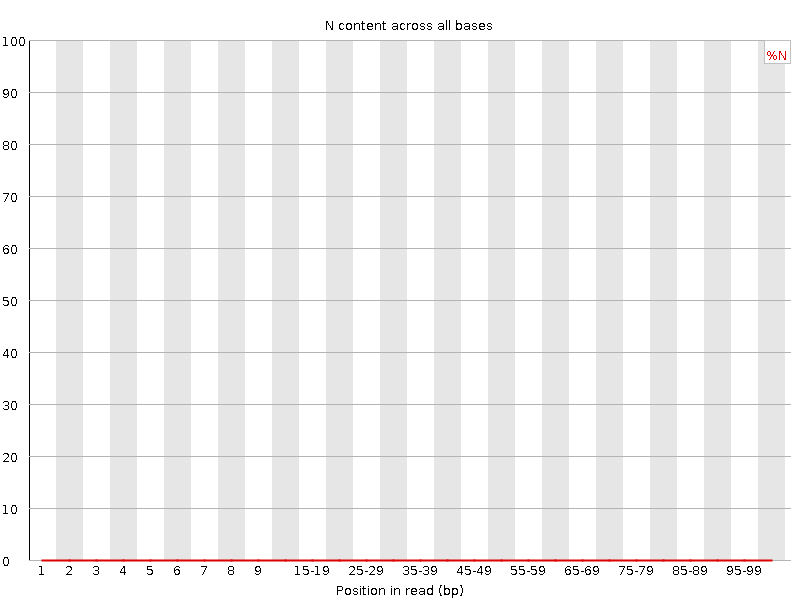
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
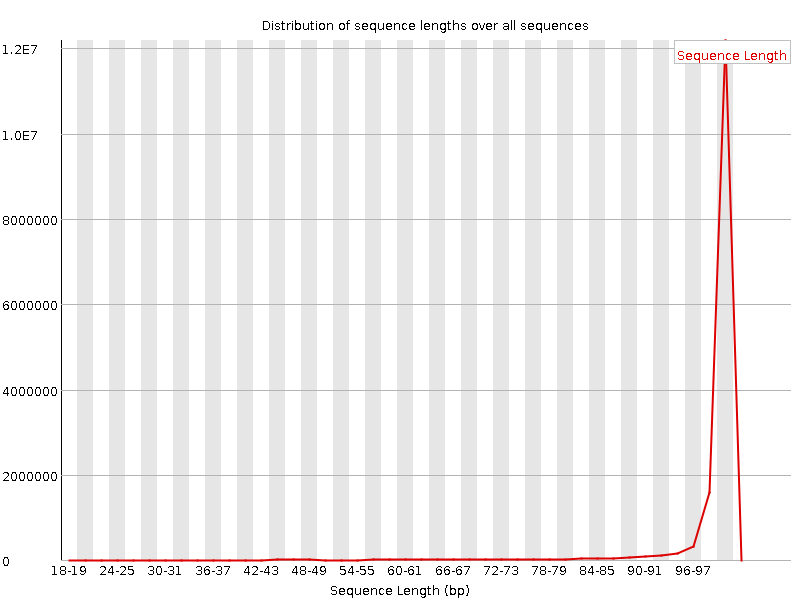
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 97491 | 0.6267707103517106 | No Hit |
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 93120 | 0.5986695033177554 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 63638 | 0.40912940133306824 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 55039 | 0.3538463358366188 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 36703 | 0.23596399033796797 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 35457 | 0.22795344264537856 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 29451 | 0.18934080264402078 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 27497 | 0.17677851517105156 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 22553 | 0.1449934848402635 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 22115 | 0.14217757802697767 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 21798 | 0.1401395815433895 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 20468 | 0.13158899692770423 | No Hit |
| CTTCTCGTCCCATAATACTATTTAAGTTTTTTTACTTAAAAAATAATTTA | 18969 | 0.12195190945483787 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 18920 | 0.12163688791636525 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 18243 | 0.11728444747665177 | No Hit |
| TTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCTT | 18206 | 0.11704657407005 | No Hit |
| CTTTGCACAGTCATTATACTGCGGCCATTTAAAAATCTCATGGGGCAGAA | 17657 | 0.11351704703695883 | No Hit |
| CCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTAAA | 17037 | 0.10953106022363188 | No Hit |
| TTATAATTAAGAAAGAATTAATTACCTTAGGGATAACAGCGTAATATTTT | 16381 | 0.10531362901469235 | No Hit |
| AATTAATTACCTTAGGGATAACAGCGTAATATTTTTAGAAAGATCATATT | 15984 | 0.10276131165196524 | No Hit |
| CAAGTTTTTAATTAAAAAACAATTGATTATGCTACCTTTGCACAGTCATT | 15968 | 0.10265844747613743 | No Hit |
| CCTCGATGTTGAATTAAAATAAAAATTAGATGTAGATGTCTAATATTTAG | 15771 | 0.10139193231125773 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content

| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1397195 | 2.9750826 | 7.2056746 | 1 |
| GCGGC | 1349910 | 2.931339 | 5.6614785 | 1 |
| CGGCG | 1144735 | 2.4858 | 9.064881 | 1 |
| CGCCG | 1143495 | 2.4348729 | 8.016288 | 1 |
| CGATC | 2498415 | 2.3589861 | 11.620287 | 4 |
| CACCG | 1667555 | 2.3517976 | 11.678848 | 1 |
| GGTCG | 1601090 | 2.3276517 | 5.8783684 | 90-94 |
| CTGGC | 1499730 | 2.1379428 | 8.365381 | 45-49 |
| CCTCG | 1505505 | 2.104486 | 6.045691 | 1 |
| GCCGG | 947065 | 2.0565584 | 5.414088 | 1 |
| CCGGC | 945365 | 2.0129895 | 6.9291477 | 1 |
| CCGAT | 2110830 | 1.993031 | 10.111406 | 3 |
| CGACG | 1367140 | 1.96631 | 5.4389234 | 1 |
| GGCTT | 1979260 | 1.8889779 | 5.6286054 | 3 |
| CGCGG | 846860 | 1.8389624 | 5.832514 | 1 |
| CCGCG | 856760 | 1.8243207 | 5.895915 | 1 |
| ACCGA | 1886695 | 1.7972912 | 7.9898596 | 2 |
| GCCGA | 1243575 | 1.7885908 | 5.5569267 | 1 |
| GACCT | 1845310 | 1.7423289 | 7.425095 | 95-97 |
| TGGCT | 1825000 | 1.7417543 | 5.9405093 | 45-49 |
| CAGCG | 1125640 | 1.618969 | 5.8037214 | 1 |
| GTCGC | 1104470 | 1.5744792 | 6.7174563 | 95-97 |
| GGGGC | 707540 | 1.5668647 | 8.36853 | 95-97 |
| GATCT | 2454970 | 1.5518438 | 7.5701456 | 5 |
| CTTTC | 2415895 | 1.4842417 | 8.196767 | 4 |
| TCCTT | 2396010 | 1.4720252 | 7.976069 | 2 |
| CGAAC | 1534845 | 1.4621142 | 5.8221107 | 90-94 |
| CTAAA | 3236240 | 1.3670241 | 6.04115 | 8 |
| CCTTT | 2196090 | 1.3492012 | 7.9683614 | 3 |
| CAAAT | 3067725 | 1.2958416 | 5.2934227 | 9 |
| CTCGG | 868350 | 1.2378781 | 5.2892594 | 1 |
| AGACC | 1266265 | 1.2062612 | 6.2103963 | 95-97 |
| CAGAC | 1250770 | 1.1915004 | 5.8447146 | 95-97 |
| TTTCG | 1823965 | 1.142779 | 8.2056465 | 5 |
| TTCGT | 1823170 | 1.1422809 | 8.247471 | 6 |
| CGTAC | 1203235 | 1.1360861 | 11.152853 | 8 |
| CTCCC | 711290 | 0.97497004 | 5.032428 | 1 |
| GTCCT | 1041740 | 0.97490907 | 10.913991 | 1 |
| TCGTA | 1472885 | 0.9310448 | 8.208048 | 7 |
| GTACA | 1419805 | 0.90549594 | 7.553194 | 9 |
| TACAA | 1959510 | 0.82771915 | 5.411053 | 7 |