![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | A22-38-R2_val_2.fq |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 19235179 |
| Filtered Sequences | 0 |
| Sequence length | 20-101 |
| %GC | 40 |
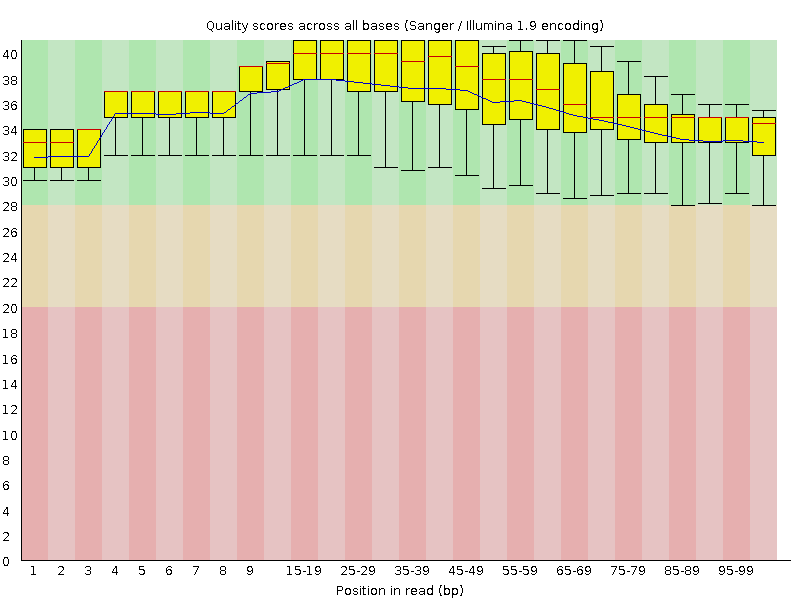
![[OK]](Icons/tick.png) Per base sequence quality
Per base sequence quality
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores

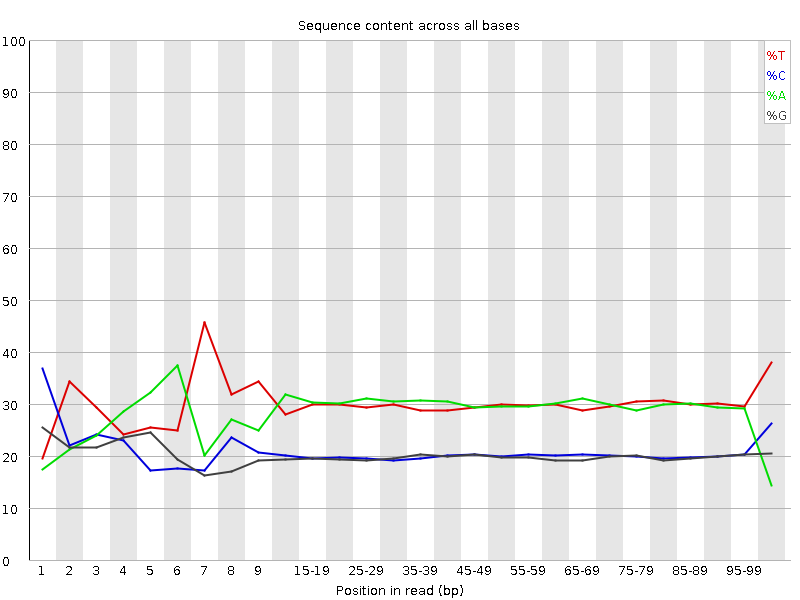
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
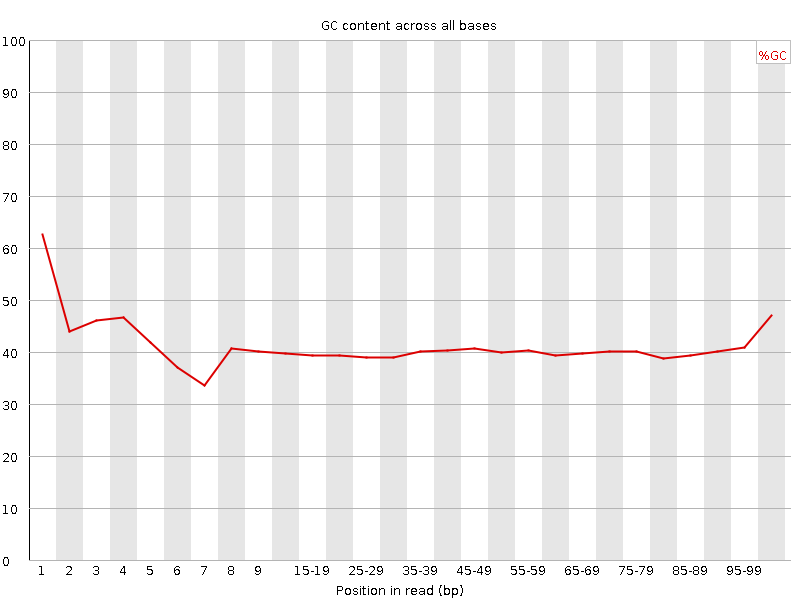
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
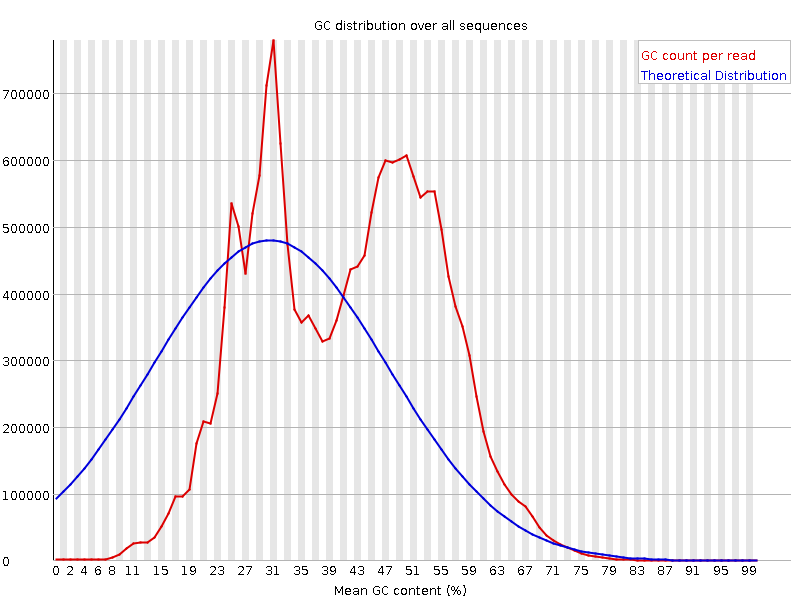
![[FAIL]](Icons/error.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content

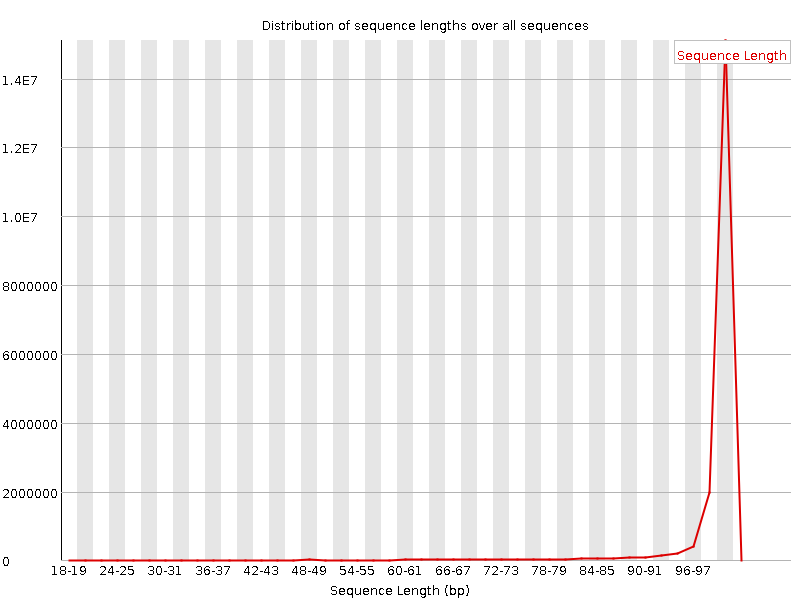
![[WARN]](Icons/warning.png) Sequence Length Distribution
Sequence Length Distribution
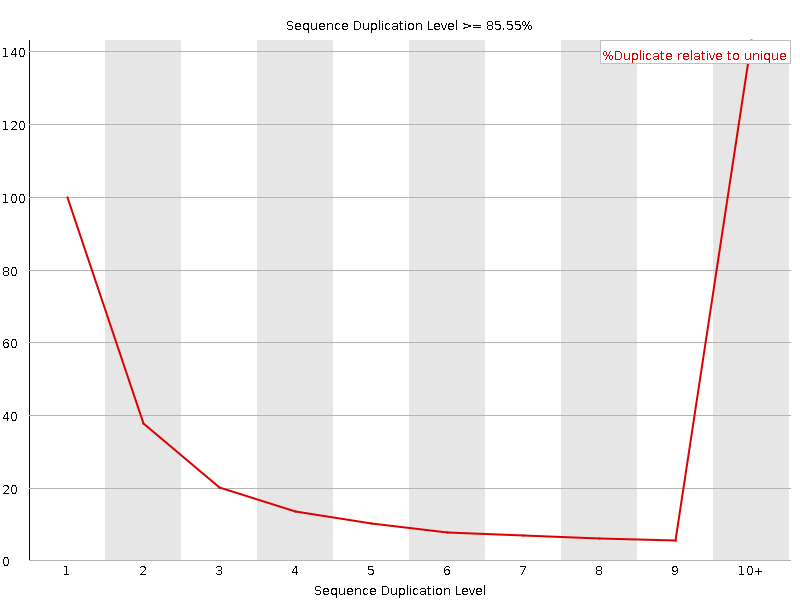
![[FAIL]](Icons/error.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GTCCTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTG | 142028 | 0.7383762844109742 | No Hit |
| CTTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCT | 126149 | 0.6558244142152251 | No Hit |
| CACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTA | 89114 | 0.46328656468442536 | No Hit |
| CTCATATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGC | 64599 | 0.3358377897081176 | No Hit |
| GCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGA | 49296 | 0.2562804328465048 | No Hit |
| CTGGCTTACACCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAA | 48329 | 0.25125318563450855 | No Hit |
| CAGCGTAATATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTC | 40759 | 0.21189821004525095 | No Hit |
| ATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAAC | 35930 | 0.18679316683249997 | No Hit |
| ATTTTTAGAAAGATCATATTAATAAAAATGATTGCGACCTCGATGTTGAA | 29986 | 0.15589145284273154 | No Hit |
| TATTAATAAAAATGATTGCGACCTCGATGTTGAATTAAAATAAAAATTAG | 29313 | 0.152392655145034 | No Hit |
| CAAATATTGAGCTCAACTGTTTATTAAAAACATAGCTTTTAGATTATAAT | 27715 | 0.14408496016595426 | No Hit |
| CCTCGATGTTGAATTAAAATAAAAATTAGATGTAGATGTCTAATATTTAG | 24021 | 0.12488056388765606 | No Hit |
| TATTAATTTAAGGAATTAGGCAAATATTGAGCTCAACTGTTTATTAAAAA | 23389 | 0.12159491731270086 | No Hit |
| CCGATCTAAACTCAAATCATGTAAAATTTTAAAGGTCGAACAGACCTAAA | 22899 | 0.11904750145553625 | No Hit |
| AATTAATTACCTTAGGGATAACAGCGTAATATTTTTAGAAAGATCATATT | 21938 | 0.1140514470907705 | No Hit |
| TTTCGTACAAATAATTTAATACTAATTATAGATAGAAACCGATCTGGCTT | 21886 | 0.11378110908143875 | No Hit |
| ATTTTCTTTCATCCAAGTTTTTAATTAAAAAACAATTGATTATGCTACCT | 21493 | 0.11173797758783528 | No Hit |
| AATTAATAAAAGGAGTAATATTAATTTAAGGAATTAGGCAAATATTGAGC | 20794 | 0.10810401088547188 | No Hit |
| TTATAATTAAGAAAGAATTAATTACCTTAGGGATAACAGCGTAATATTTT | 20772 | 0.10798963711229305 | No Hit |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
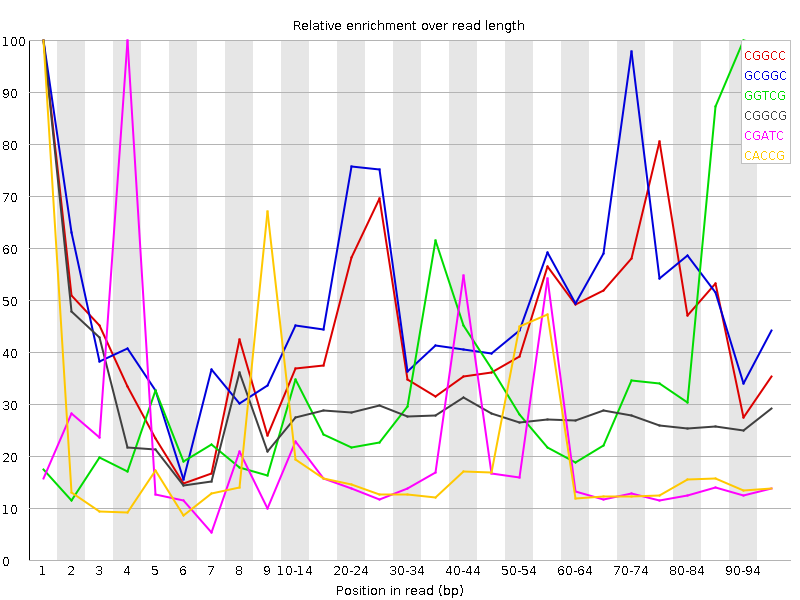
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| CGGCC | 1789300 | 2.9226892 | 6.3152275 | 1 |
| GCGGC | 1694185 | 2.823181 | 5.397974 | 1 |
| GGTCG | 2143515 | 2.435492 | 6.4505806 | 90-94 |
| CGGCG | 1461075 | 2.4347277 | 8.534476 | 1 |
| CGATC | 3241860 | 2.4305496 | 12.302326 | 4 |
| CACCG | 2189885 | 2.40796 | 12.629421 | 1 |
| CGCCG | 1460915 | 2.3862965 | 7.7930474 | 1 |
| CTGGC | 1998100 | 2.2253537 | 9.104851 | 45-49 |
| CCTCG | 1979975 | 2.1615393 | 6.5389085 | 1 |
| GCCGG | 1286220 | 2.1433501 | 5.1954427 | 1 |
| CCGGC | 1278010 | 2.087535 | 6.991285 | 1 |
| CCGAT | 2756550 | 2.066694 | 10.776421 | 3 |
| GGCTT | 2542110 | 1.9304495 | 5.8534307 | 3 |
| CGACG | 1715695 | 1.9246265 | 5.0508723 | 1 |
| ACCGA | 2471005 | 1.8659863 | 8.645701 | 2 |
| CGCGG | 1102455 | 1.8371252 | 5.499631 | 1 |
| GACCT | 2437795 | 1.8277109 | 7.840917 | 95-97 |
| CCGCG | 1116715 | 1.8240716 | 5.4529114 | 1 |
| AGGTC | 2382165 | 1.8220512 | 5.659388 | 95-97 |
| TGGCT | 2363170 | 1.7945645 | 6.471436 | 45-49 |
| GCCGA | 1564030 | 1.7544922 | 5.409355 | 1 |
| GATCT | 3204740 | 1.6382699 | 8.222689 | 5 |
| CAGCG | 1433840 | 1.6084481 | 6.373732 | 1 |
| GTCGC | 1416035 | 1.5770875 | 7.253781 | 95-97 |
| CGAAC | 2001205 | 1.5112156 | 6.3260865 | 90-94 |
| TCCTT | 3000510 | 1.4927418 | 9.041921 | 2 |
| CTTTC | 2980720 | 1.4828964 | 9.099937 | 4 |
| GGGGC | 867820 | 1.4753191 | 7.8939667 | 95-97 |
| CTAAA | 4088220 | 1.4170326 | 6.418431 | 8 |
| CCTTT | 2718515 | 1.3524506 | 8.924765 | 3 |
| CAAAT | 3840920 | 1.3313152 | 5.777865 | 9 |
| AGACC | 1650420 | 1.2463193 | 6.6045656 | 95-97 |
| ATCTA | 3556235 | 1.223803 | 5.0356207 | 6 |
| CAGAC | 1607610 | 1.2139912 | 6.31695 | 95-97 |
| CTCGG | 1079020 | 1.2017422 | 5.2132645 | 1 |
| CGTAC | 1582660 | 1.1865823 | 12.571984 | 8 |
| TTCGT | 2313270 | 1.1740708 | 9.285786 | 6 |
| TTTCG | 2297810 | 1.1662245 | 9.258261 | 5 |
| GTCCT | 1308415 | 0.9739378 | 12.339434 | 1 |
| TCGTA | 1901480 | 0.9720404 | 9.288748 | 7 |
| GTACA | 1804740 | 0.9292484 | 8.632555 | 9 |
| ACAAA | 2572650 | 0.89815414 | 5.4259725 | 8 |
| TACAA | 2394930 | 0.83011544 | 5.904102 | 7 |