Wednesday notes
Open science
Another pre-eminent scholar that signs reviews
Human genetics
Some ideas for a ‘motivating example’ for the “Alleles changing” lecture I will be covering in Amanda Yonin’s Human Genetics course on Oct 25th.
Combine this paper on rapid increase in rare variants with a discussion of Mendelian disease in this population using this case study
Accelerating selection in humans
ApTranscriptome
Turning sim-transcriptome.sh into a script that can be run with fasta file of known transcripts as input, and output including
- assembly stats for all reads
- assembly stats for normalized reads
- gene expression counts of reads on known transcripts
- gene expression counts of reads on assembled transcripts
Assembly
Surprising result that velvet-oases assembly with all reads results in fewer contigs of shorter length than assembly with normalized reads. Why would this be???
filename sim-oases-21/transcripts.fa
| assembly stat | result |
|---|---|
| Total Contigs | 201 |
| Total Trimmed Contigs | 201 |
| Total Length | 63728 |
| Min contig size | 106 |
| Median contig size | 298 |
| Mean contig size | 317 |
| Max contig size | 686 |
| N50 Contig | 65 |
| N50 Length | 390 |
| N90 Contig | 159 |
| N90 Length | 183 |
filename sim-oases-norm-21/transcripts.fa
| assembly stat | result |
|---|---|
| Total Contigs | 322 |
| Total Trimmed Contigs | 322 |
| Total Length | 128575 |
| Min contig size | 101 |
| Median contig size | 416 |
| Mean contig size | 399 |
| Max contig size | 629 |
| N50 Contig | 123 |
| N50 Length | 462 |
| N90 Contig | 257 |
| N90 Length | 281 |
Gene Expression
Tophat-cufflinks
Parial explanation to (problem before)(/aptranscriptome/2013/10/09/more-simulation-notes.html) where only ~29 of 100 transcripts actually had gene expression counts - low read mapping to assembled transcripts.
- map reads to known transcripts
- FPKM for 97 transcripts
- map reads to assembled transcripts
- FPKM for 31 transcripts
Could be because of lots of multiple alignment to the assembled transcripts vs the known transcripts.
For tophat mapped reads to assembled transcripts:
> Mapped: 288471 (19.4% of input)
> of these: 183776 (63.7%) have multiple alignments (0 have >20)
> Right reads:
> Input: 1488635
> Mapped: 276348 (18.6% of input)
> of these: 176708 (63.9%) have multiple alignments (0 have >20)
> 19.0% overall read alignment rate.
>
> Aligned pairs: 186284
> of these: 110648 (59.4%) have multiple alignments
> and: 2 ( 0.0%) are discordant alignments
> 12.5% concordant pair alignment rate.compared to tophat mapped reads to known transcripts
> Left reads:
> Input: 1488635
> Mapped: 978717 (65.7% of input)
> of these: 78299 ( 8.0%) have multiple alignments (0 have >20)
> Right reads:
> Input: 1488635
> Mapped: 973857 (65.4% of input)
> of these: 76004 ( 7.8%) have multiple alignments (0 have >20)
> 65.6% overall read alignment rate.
>
> Aligned pairs: 964826
> of these: 62730 ( 6.5%) have multiple alignments
> and: 3 ( 0.0%) are discordant alignments
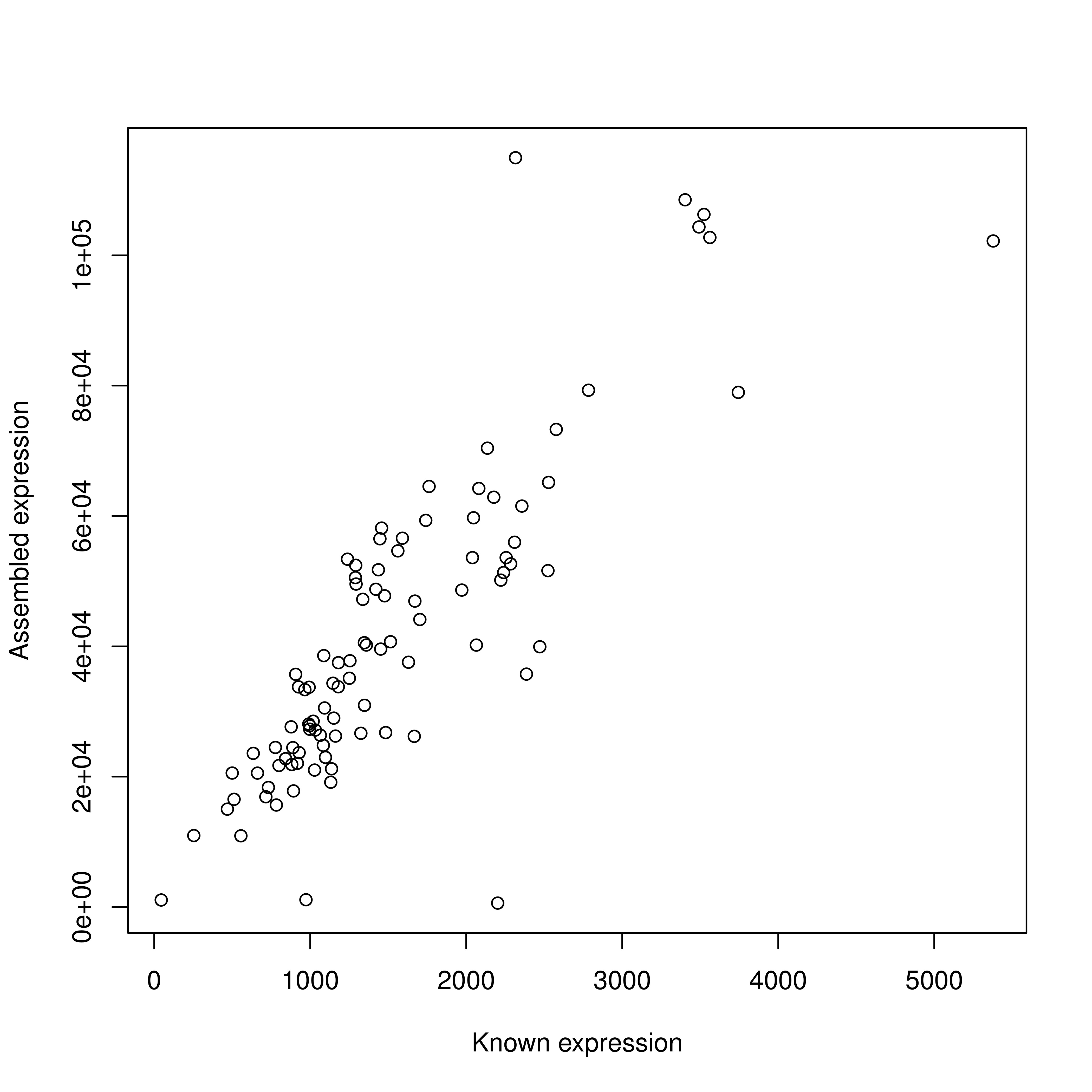
> 64.8% concordant pair alignment rate.But, the correlation of expression counts from cufflinks mapped to known transcripts is beautiful! r=0.83 for 97 of 100 transcripts!
Known vs assembled transcript expression
Similar problem with BWA…what to do with real data where I can’t infer incorrect isoforms?
