ddRADseq debugging
Pooling and size selection
- Pooled all remaining ligated sample without AMPure bead purification
- Ran on 1.5% agarose gel for 3 hours
- still no DNA visible in sample lane when I visualize gel
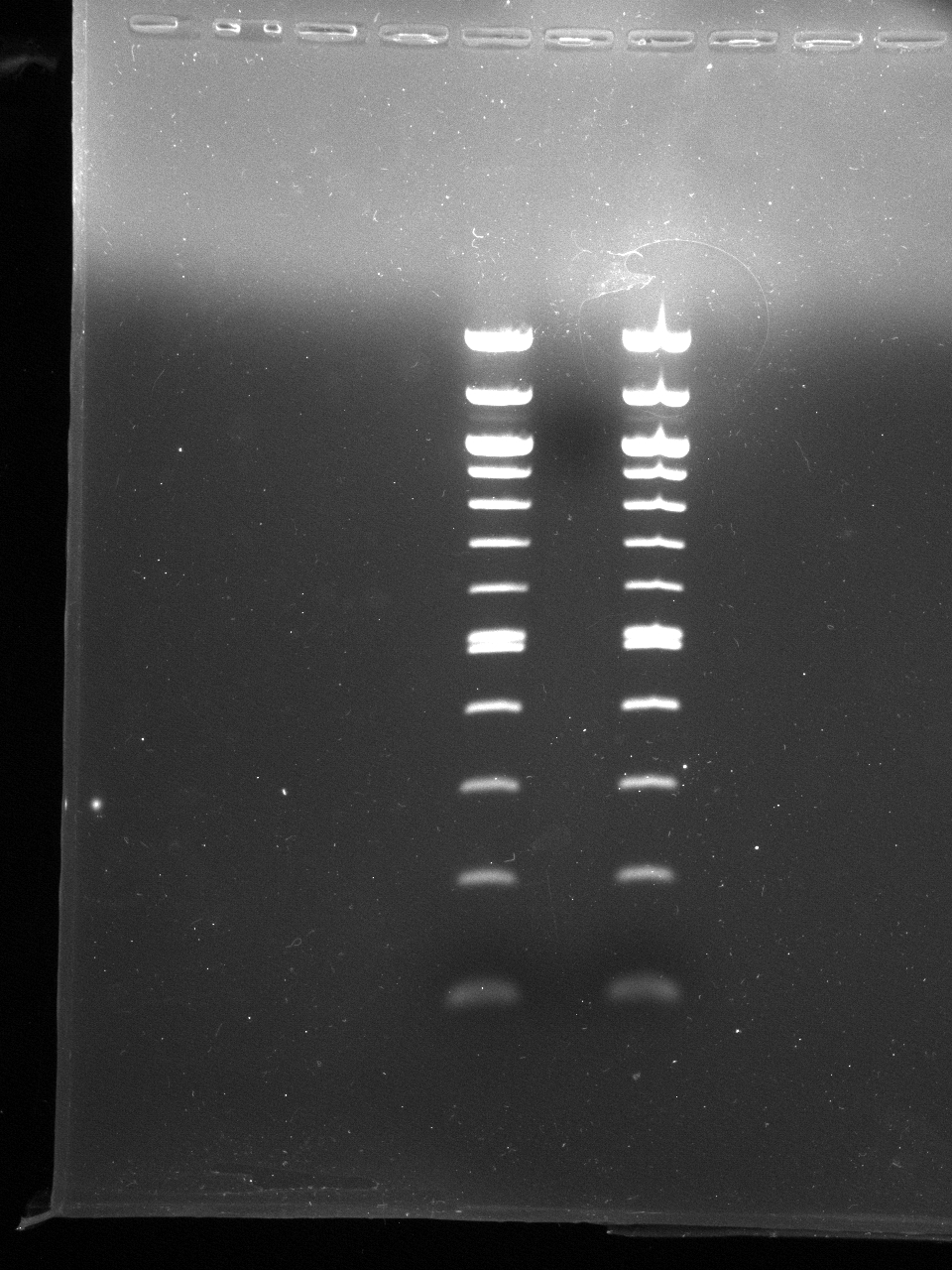
- Excised gel fragments for 275, 375 and 475 bp +- 25bp
- QIAquick gel purification
DNA concentration from Qubit
| Sample | Stock conc (ng/ul) | Total DNA (ng) |
|---|---|---|
| P | 8.78 | 351.2 |
| P-200 | 0.055 | 0.5 |
| P-300 | 0.066 | 0.6 |
| P-400 | 0 | 0 |
Total DNA is in 40 ul for sample P and 10 ul for P-200, P-300 and P-400
Key points:
- DNA is lost during AMPure purification step during pooling
- From Bioanalyzer, expect about 10% of DNA to be in the 200 bp window, so could maximally recover about 8 ng, yet recover less than 1. So, even the gel purification is problematic.
Seems that I need more input DNA…though I’m using within recommended range (100 - 500 ng) from ddRADseq protocol. What gives??
ApTranscriptome
Writing…
Silene
Phone call with SK - discussed analyses to finish Silene ms
