Aphaenogaster QC report for Illumina
Update from Minnesota BMGC with RNA QC report and Agilent profiles for the 24 Aphaenogaster RNA samples submitted for Illumina NGS project ‘Stanton-Geddes_Project_001’
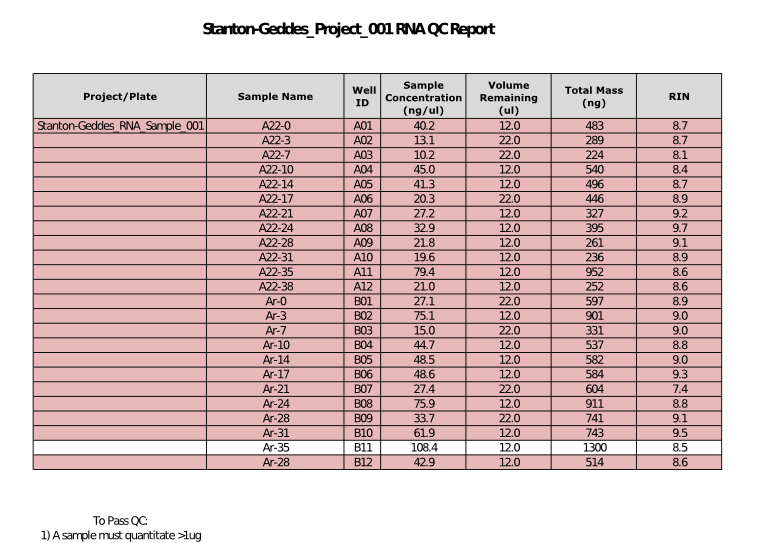
Results:
As expected, many of these samples are failing QC for mass. Quality looks excellent though. It is typical for insect RNA to not segregate into 18S and 28S bands. The baseline is noise free indicating the RNA is in good shape.
If you are unable to easily obtain additional sample (as you suggested would be the case) I think the approach we should take for this experiment is to attempt library preparation with 200ng of input total RNA rather the the standard low input amount of 500ng. Based on previous experience, this should produce sufficient library to pool and sequence.Based on recommendation and other reading that samples down to 100ng still give good sequencing results on Illumina HiSeq, gave go ahead for sequencing!
QC Result: